- Record: found
- Abstract: found
- Article: found
Dual function of GTPBP6 in biogenesis and recycling of human mitochondrial ribosomes
Read this article at
Abstract
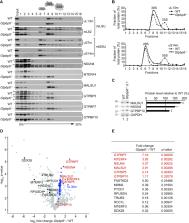
Translation and ribosome biogenesis in mitochondria require auxiliary factors that ensure rapid and accurate synthesis of mitochondrial proteins. Defects in translation are associated with oxidative phosphorylation deficiency and cause severe human diseases, but the exact roles of mitochondrial translation-associated factors are not known. Here we identify the functions of GTPBP6, a homolog of the bacterial ribosome-recycling factor HflX, in human mitochondria. Similarly to HflX, GTPBP6 facilitates the dissociation of ribosomes in vitro and in vivo. In contrast to HflX, GTPBP6 is also required for the assembly of mitochondrial ribosomes. GTPBP6 ablation leads to accumulation of late assembly intermediate(s) of the large ribosomal subunit containing ribosome biogenesis factors MTERF4, NSUN4, MALSU1 and the GTPases GTPBP5, GTPBP7 and GTPBP10. Our data show that GTPBP6 has a dual function acting in ribosome recycling and biogenesis. These findings contribute to our understanding of large ribosomal subunit assembly as well as ribosome recycling pathway in mitochondria.
Related collections
Most cited references66
- Record: found
- Abstract: found
- Article: not found
MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification.
- Record: found
- Abstract: found
- Article: not found
The Perseus computational platform for comprehensive analysis of (prote)omics data.
- Record: found
- Abstract: found
- Article: not found