- Record: found
- Abstract: found
- Article: found
Reprogramming of T cell‐derived small extracellular vesicles using IL2 surface engineering induces potent anti‐cancer effects through miRNA delivery
Read this article at
Abstract
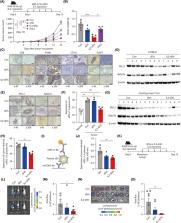
T cell‐derived small extracellular vesicles (sEVs) exhibit anti‐cancer effects. However, their anti‐cancer potential should be reinforced to enhance clinical applicability. Herein, we generated interleukin‐2‐tethered sEVs (IL2‐sEVs) from engineered Jurkat T cells expressing IL2 at the plasma membrane via a flexible linker to induce an autocrine effect. IL2‐sEVs increased the anti‐cancer ability of CD8 + T cells without affecting regulatory T (T reg) cells and down‐regulated cellular and exosomal PD‐L1 expression in melanoma cells, causing their increased sensitivity to CD8 + T cell‐mediated cytotoxicity. Its effect on CD8 + T and melanoma cells was mediated by several IL2‐sEV‐resident microRNAs (miRNAs), whose expressions were upregulated by the autocrine effects of IL2. Among the miRNAs, miR‐181a‐3p and miR‐223‐3p notably reduced the PD‐L1 protein levels in melanoma cells. Interestingly, miR‐181a‐3p increased the activity of CD8 + T cells while suppressing T reg cell activity. IL2‐sEVs inhibited tumour progression in melanoma‐bearing immunocompetent mice, but not in immunodeficient mice. The combination of IL2‐sEVs and existing anti‐cancer drugs significantly improved anti‐cancer efficacy by decreasing PD‐L1 expression in vivo. Thus, IL2‐sEVs are potential cancer immunotherapeutic agents that regulate both immune and cancer cells by reprogramming miRNA levels.
Related collections
Most cited references72
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found
edgeR: a Bioconductor package for differential expression analysis of digital gene expression data

- Record: found
- Abstract: found
- Article: found