- Record: found
- Abstract: found
- Article: found
New Insights in the Removal of the Hydantoins, Oxidation Product of Pyrimidines, via the Base Excision and Nucleotide Incision Repair Pathways
Read this article at
Abstract
Background
Oxidative damage to DNA, if not repaired, can be both miscoding and blocking. These genetic alterations can lead to mutations and/or cell death, which in turn cause cancer and aging. Oxidized DNA bases are substrates for two overlapping repair pathways: base excision (BER) and nucleotide incision repair (NIR). Hydantoin derivatives such as 5-hydroxyhydantoin (5OH-Hyd) and 5-methyl-5-hydroxyhydantoin (5OH-5Me-Hyd), major products of cytosine and thymine oxidative degradation pathways, respectively, have been detected in cancer cells and ancient DNA. Hydantoins are blocking lesions for DNA polymerases and excised by bacterial and yeast DNA glycosylases in the BER pathway. However little is known about repair of pyrimidine-derived hydantoins in human cells.
Methodology/Principal Findings
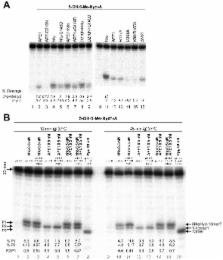
Here, using both denaturing PAGE and MALDI-TOF MS analyses we report that the bacterial, yeast and human AP endonucleases can incise duplex DNA 5′ next to 5OH-Hyd and 5OH-5Me-Hyd thus initiating the NIR pathway. We have fully reconstituted the NIR pathway for these lesions in vitro using purified human proteins. Depletion of Nfo in E. coli and APE1 in HeLa cells abolishes the NIR activity in cell-free extracts. Importantly, a number of redundant DNA glycosylase activities can excise hydantoin residues, including human NTH1, NEIL1 and NEIL2 and the former protein being a major DNA glycosylase activity in HeLa cells extracts.
Related collections
Most cited references68

- Record: found
- Abstract: found
- Article: found
Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA
- Record: found
- Abstract: not found
- Article: not found
The biochemical basis of zinc physiology.
- Record: found
- Abstract: not found
- Article: not found