- Record: found
- Abstract: found
- Article: found
Leafcutter ants enhance microbial drought resilience in tropical forest soil
Read this article at
Abstract
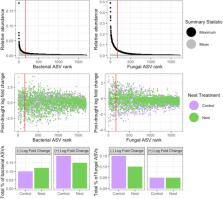
We conducted a research campaign in a neotropical rainforest in Costa Rica throughout the drought phase of an El‐Nino Southern Oscillation event to determine microbial community dynamics and soil C fluxes. Our study included nests of the leafcutter ant Atta cephalotes, as soil disturbances made by these ecosystem engineers may influence microbial drought response. Drought decreased the diversity of microbes and the abundance of core microbiome taxa, including Verrucomicrobial bacteria and Sordariomycete fungi. Despite initial responses of decreasing diversity and altered composition, 6 months post‐drought the microbiomes were similar to pre‐drought conditions, demonstrating the resilience of soil microbial communities to drought events. A. cephalotes nests altered fungal composition in the surrounding soil, and reduced both fungal mortality and growth of Acidobacteria post‐drought. Drought increased CH 4 consumption in soils due to lower soil moisture, and A. cephalotes nests decrease the variability of CH 4 emissions in some soil types. CH 4 emissions were tracked by the abundance of methanotrophic bacteria and fungal composition. These results characterize the microbiome of tropical soils across both time and space during drought and provide evidence for the importance of leafcutter ant nests in shaping soil microbiomes and enhancing microbial resilience during climatic perturbations.
Abstract
We carried out an eight‐month investigation on a tropical soil microbial community in the aftermath of a severe drought event, which resulted in changes to soil moisture levels and methane emissions. Our study reveals that the leafcutter ant, Atta cephalotes, plays a crucial role in stabilizing the microbial community and enhancing its resilience by mitigating the mortality rates of fungi and methanotrophs. The findings underscore the significance of multi‐trophic interactions in bolstering ecosystem resilience in the face of extreme climatic occurrences.
Related collections
Most cited references48

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: not found
- Article: not found
Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2

- Record: found
- Abstract: found
- Article: found
