- Record: found
- Abstract: found
- Article: found
Three-Fingered RAVERs: Rapid Accumulation of Variations in Exposed Residues of Snake Venom Toxins
Read this article at
Abstract
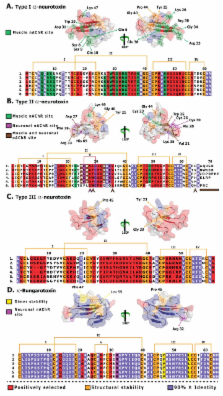
Three-finger toxins (3FTx) represent one of the most abundantly secreted and potently toxic components of colubrid (Colubridae), elapid (Elapidae) and psammophid (Psammophiinae subfamily of the Lamprophidae) snake venom arsenal. Despite their conserved structural similarity, they perform a diversity of biological functions. Although they are theorised to undergo adaptive evolution, the underlying diversification mechanisms remain elusive. Here, we report the molecular evolution of different 3FTx functional forms and show that positively selected point mutations have driven the rapid evolution and diversification of 3FTx. These diversification events not only correlate with the evolution of advanced venom delivery systems (VDS) in Caenophidia, but in particular the explosive diversification of the clade subsequent to the evolution of a high pressure, hollow-fanged VDS in elapids, highlighting the significant role of these toxins in the evolution of advanced snakes. We show that Type I, II and III α-neurotoxins have evolved with extreme rapidity under the influence of positive selection. We also show that novel Oxyuranus/ Pseudonaja Type II forms lacking the apotypic loop-2 stabilising cysteine doublet characteristic of Type II forms are not phylogenetically basal in relation to other Type IIs as previously thought, but are the result of secondary loss of these apotypic cysteines on at least three separate occasions. Not all 3FTxs have evolved rapidly: κ-neurotoxins, which form non-covalently associated heterodimers, have experienced a relatively weaker influence of diversifying selection; while cytotoxic 3FTx, with their functional sites, dispersed over 40% of the molecular surface, have been extremely constrained by negative selection. We show that the a previous theory of 3FTx molecular evolution (termed ASSET) is evolutionarily implausible and cannot account for the considerable variation observed in very short segments of 3FTx. Instead, we propose a theory of Rapid Accumulation of Variations in Exposed Residues (RAVER) to illustrate the significance of point mutations, guided by focal mutagenesis and positive selection in the evolution and diversification of 3FTx.
Related collections
Most cited references111
- Record: found
- Abstract: found
- Article: not found
A codon-based model of nucleotide substitution for protein-coding DNA sequences.
- Record: found
- Abstract: found
- Article: not found
Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology.
- Record: found
- Abstract: found
- Article: not found