- Record: found
- Abstract: found
- Article: found
Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system
Read this article at
Abstract
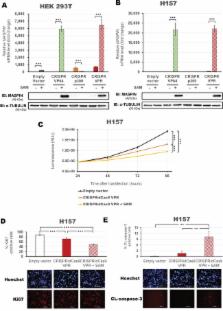
The aberrant epigenetic silencing of tumor suppressor genes (TSGs) plays a major role during carcinogenesis and regaining these dormant functions by engineering of sequence-specific epigenome editing tools offers a unique opportunity for targeted therapies. However, effectively normalizing the expression and regaining tumor suppressive functions of silenced TSGs by artificial transcription factors (ATFs) still remains a major challenge. Herein we describe novel combinatorial strategies for the potent reactivation of two class II TSGs, MASPIN and REPRIMO, in cell lines with varying epigenetic states, using the CRISPR/dCas9 associated system linked to a panel of effector domains (VP64, p300, VPR and SAM complex), as well as with protein-based ATFs, Zinc Fingers and TALEs. We found that co-delivery of multiple effector domains using a combination of CRISPR/dCas9 and TALEs or SAM complex maximized activation in highly methylated promoters. In particular, CRISPR/dCas9 VPR with SAM upregulated MASPIN mRNA (22,145-fold change) in H157 lung cancer cells, with accompanying re-expression of MASPIN protein, which led to a concomitant inhibition of cell proliferation and induction of apoptotic cell death. Consistently, CRISPR/dCas9 VP64 with SAM upregulated REPRIMO (680-fold change), which led to phenotypic reprogramming in AGS gastric cancer cells. Altogether, our results outlined novel sequence-specific, combinatorial epigenome editing approaches to reactivate highly methylated TSGs as a promising therapy for cancer and other diseases.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression.
- Record: found
- Abstract: found
- Article: not found
Epigenome editing by a CRISPR/Cas9-based acetyltransferase activates genes from promoters and enhancers
- Record: found
- Abstract: found
- Article: not found