- Record: found
- Abstract: found
- Article: found
SppI Forms a Membrane Protein Complex with SppA and Inhibits Its Protease Activity in Bacillus subtilis
Read this article at
Abstract
Our study presents new insights into the molecular mechanism that regulates the activity of SppA, a widely conserved bacterial membrane protease. We show that the membrane proteins SppA and SppI form a complex in the Gram-positive model bacterium B. subtilis and that SppI inhibits SppA protease activity in vitro and in vivo. Furthermore, we demonstrate that the C-terminal domain of SppI is involved in SppA inhibition. Since SppA, through its protease activity, contributes directly to resistance to lantibiotic peptides and cationic antibacterial peptides, we propose that the conserved SppA-SppI complex could play a major role in the evasion of bactericidal peptides, including those produced as part of human innate immune defenses.
ABSTRACT
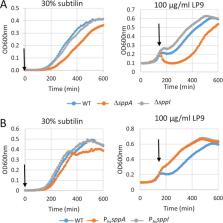
The membrane protease SppA of Bacillus subtilis was first described as a signal peptide peptidase and later shown to confer resistance to lantibiotics. Here, we report that SppA forms octameric complexes with YteJ, a membrane protein of thus-far-unknown function. Interestingly, sppA and yteJ deletion mutants exhibited no protein secretion defects. However, these mutant strains differed significantly in their resistance to antimicrobial peptides. In particular, sppA mutant cells displayed increased sensitivity to the lantibiotics nisin and subtilin and the human lysozyme-derived cationic antimicrobial peptide LP9. Importantly, YteJ was shown to antagonize SppA activity both in vivo and in vitro, and this SppA-inhibitory activity involved the C-terminal domain of YteJ, which was therefore renamed SppI. Most likely, SppI-mediated control is needed to protect B. subtilis against the potentially detrimental protease activity of SppA since a mutant overexpressing sppA by itself displayed defects in cell division. Altogether, we conclude that the SppA-SppI complex of B. subtilis has a major role in protection against antimicrobial peptides.
IMPORTANCE Our study presents new insights into the molecular mechanism that regulates the activity of SppA, a widely conserved bacterial membrane protease. We show that the membrane proteins SppA and SppI form a complex in the Gram-positive model bacterium B. subtilis and that SppI inhibits SppA protease activity in vitro and in vivo. Furthermore, we demonstrate that the C-terminal domain of SppI is involved in SppA inhibition. Since SppA, through its protease activity, contributes directly to resistance to lantibiotic peptides and cationic antibacterial peptides, we propose that the conserved SppA-SppI complex could play a major role in the evasion of bactericidal peptides, including those produced as part of human innate immune defenses.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein.
- Record: found
- Abstract: found
- Article: not found
Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis.
- Record: found
- Abstract: found
- Article: not found