- Record: found
- Abstract: found
- Article: found
Prediction of Pseudoprogression versus Progression using Machine Learning Algorithm in Glioblastoma
Read this article at
Abstract
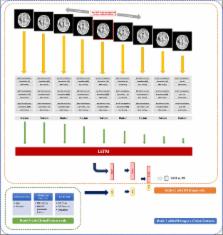
We aimed to investigate the feasibility of machine learning (ML) algorithm to distinguish pseudoprogression (PsPD) from progression (PD) in patients with glioblastoma (GBM). We recruited the patients diagnosed as primary GBM who received gross total resection (GTR) and concurrent chemoradiotherapy in two institutions from April 2010 to April 2017 and presented suspicious contrast-enhanced lesion on brain magnetic resonance imaging (MRI) during follow-up. Patients from two institutions were allocated to training (N = 59) and testing (N = 19) datasets, respectively. We developed a convolutional neural network combined with a long short-term memory ML structure. MRI data, which was 9 axial post-contrast T1-weighted images in our study, and clinical features were incorporated (Model 1). In the testing set, the trained Model 1 resulted in AUC of 0.83, AUPRC of 0.87, and F1-score of 0.74 using optimal threshold. The performance was superior to that of Model 2 (CNN-LSTM model with MRI data alone) and Model 3 (random forest model with clinical feature alone). The developed algorithm involving MRI data and clinical features could help making decision during follow-up of patients with GBM treated with GTR and concurrent CCRT.
Related collections
Most cited references22
- Record: found
- Abstract: not found
- Book Chapter: not found
Mitosis Detection in Breast Cancer Histology Images with Deep Neural Networks
- Record: found
- Abstract: found
- Article: not found
MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients.
- Record: found
- Abstract: found
- Article: not found