- Record: found
- Abstract: found
- Article: found
Single-cell profiling of the human decidual immune microenvironment in patients with recurrent pregnancy loss
Read this article at
Abstract
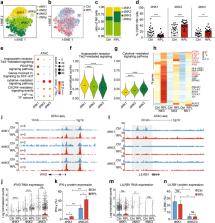
Maintaining homeostasis of the decidual immune microenvironment at the maternal–fetal interface is essential for placentation and reproductive success. Although distinct decidual immune cell subpopulations have been identified under normal conditions, systematic understanding of the spectrum and heterogeneity of leukocytes under recurrent miscarriage in human deciduas remains unclear. To address this, we profiled the respective transcriptomes of 18,646 primary human decidual immune cells isolated from patients with recurrent pregnancy loss (RPL) and healthy controls at single-cell resolution. We discovered dramatic differential distributions of immune cell subsets in RPL patients compared with the normal decidual immune microenvironment. Furthermore, we found a subset of decidual natural killer (NK) cells that support embryo growth were diminished in proportion due to abnormal NK cell development in RPL patients. We also elucidated the altered cellular interactions between the decidual immune cell subsets in the microenvironment and those of the immune cells with stromal cells and extravillous trophoblast under disease state. These results provided deeper insights into the RPL decidual immune microenvironment disorder that are potentially applicable to improve the diagnosis and therapeutics of this disease.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.

- Record: found
- Abstract: found
- Article: found
BEDTools: a flexible suite of utilities for comparing genomic features

- Record: found
- Abstract: found
- Article: found