- Record: found
- Abstract: found
- Article: found
Epigenetic Regulation of Angiogenesis in Peripheral Artery Disease
Read this article at
Abstract
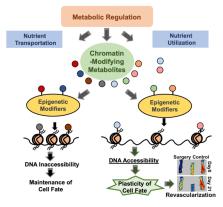
Peripheral arterial disease (PAD) represents a global health concern with a rising prevalence attributed to factors such as obesity, diabetes, aging, and smoking. Among patients with PAD, chronic limb-threatening ischemia (CLTI) is the most severe manifestation, associated with substantial morbidity and mortality. While revascularization remains the primary therapy for CLTI, not all patients are candidates for such interventions, highlighting the need for alternative approaches. Impaired angiogenesis, the growth of new blood vessels, is a central feature of PAD, and despite decades of research, effective clinical treatments remain elusive. Epigenetics, the study of heritable changes in gene expression, has gained prominence in understanding PAD pathogenesis. Here, we explore the role of epigenetic regulation in angiogenesis within the context of PAD, with a focus on long non-coding RNAs and fibroblast-endothelial cell transdifferentiation. Additionally, we discuss the interplay between metabolic control and epigenetic regulation, providing insights into potential novel therapeutic avenues for improving PAD treatments. This review aims to offer a concise update on the application of epigenetics in angiogenesis and PAD research, inspiring further investigations in this promising field.
Related collections
Most cited references78
- Record: found
- Abstract: found
- Article: not found
Functional Classification and Experimental Dissection of Long Noncoding RNAs
- Record: found
- Abstract: found
- Article: not found
Regulation of chromatin by histone modifications.
- Record: found
- Abstract: found
- Article: not found