- Record: found
- Abstract: found
- Article: found
Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes

Read this article at
Summary
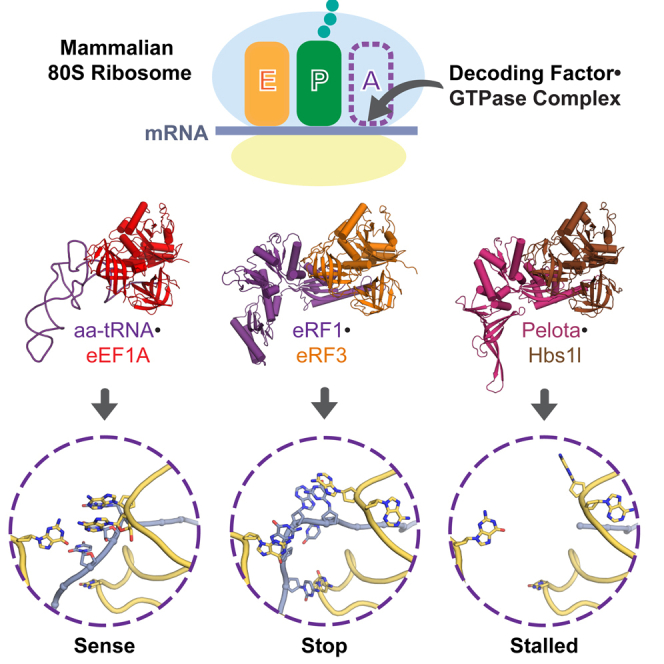
In eukaryotes, accurate protein synthesis relies on a family of translational GTPases that pair with specific decoding factors to decipher the mRNA code on ribosomes. We present structures of the mammalian ribosome engaged with decoding factor⋅GTPase complexes representing intermediates of translation elongation (aminoacyl-tRNA⋅eEF1A), termination (eRF1⋅eRF3), and ribosome rescue (Pelota⋅Hbs1l). Comparative analyses reveal that each decoding factor exploits the plasticity of the ribosomal decoding center to differentially remodel ribosomal proteins and rRNA. This leads to varying degrees of large-scale ribosome movements and implies distinct mechanisms for communicating information from the decoding center to each GTPase. Additional structural snapshots of the translation termination pathway reveal the conformational changes that choreograph the accommodation of decoding factors into the peptidyl transferase center. Our results provide a structural framework for how different states of the mammalian ribosome are selectively recognized by the appropriate decoding factor⋅GTPase complex to ensure translational fidelity.
Graphical Abstract
Highlights
-
•
Cryo-EM structures of elongating, terminating, and stalled mammalian ribosomes
-
•
Eukaryotic-specific elements contribute to stringent sense and stop codon decoding
-
•
Pelota engages stalled ribosomes by destabilizing mRNA in the mRNA channel
-
•
Decoding complexes communicate recognition to GTPase activation in different ways
Abstract
The individual decoding factor⋅GTPase complexes involved in protein synthesis differentially remodel local protein and RNA elements on ribosomes to ensure translation fidelity.
Related collections
Most cited references40

- Record: found
- Abstract: found
- Article: found
Sampling the conformational space of the catalytic subunit of human γ-secretase
- Record: found
- Abstract: found
- Article: not found
The elongation, termination, and recycling phases of translation in eukaryotes.
- Record: found
- Abstract: found
- Article: not found