- Record: found
- Abstract: found
- Article: found
Artificial intelligence in colonoscopy: from detection to diagnosis
Read this article at
Abstract
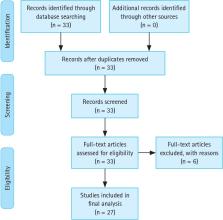
This study reviews the recent progress of artificial intelligence for colonoscopy from detection to diagnosis. The source of data was 27 original studies in PubMed. The search terms were “colonoscopy” (title) and “deep learning” (abstract). The eligibility criteria were: (1) the dependent variable of gastrointestinal disease; (2) the interventions of deep learning for classification, detection and/or segmentation for colonoscopy; (3) the outcomes of accuracy, sensitivity, specificity, area under the curve (AUC), precision, F1, intersection of union (IOU), Dice and/or inference frames per second (FPS); (3) the publication year of 2021 or later; (4) the publication language of English. Based on the results of this study, different deep learning methods would be appropriate for different tasks for colonoscopy, e.g., Efficientnet with neural architecture search (AUC 99.8%) in the case of classification, You Only Look Once with the instance tracking head (F1 96.3%) in the case of detection, and Unet with dense-dilation-residual blocks (Dice 97.3%) in the case of segmentation. Their performance measures reported varied within 74.0–95.0% for accuracy, 60.0–93.0% for sensitivity, 60.0–100.0% for specificity, 71.0–99.8% for the AUC, 70.1–93.3% for precision, 81.0–96.3% for F1, 57.2–89.5% for the IOU, 75.1–97.3% for Dice and 66–182 for FPS. In conclusion, artificial intelligence provides an effective, non-invasive decision support system for colonoscopy from detection to diagnosis.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
Mastering the game of Go with deep neural networks and tree search.
- Record: found
- Abstract: found
- Article: not found
U-Net: Convolutional Networks for Biomedical Image Segmentation
- Record: found
- Abstract: found
- Article: not found