- Record: found
- Abstract: found
- Article: found
Classification of arrhythmias using an LSTM- and GAN-based approach to ECG signal augmentation
Read this article at
Abstract
Funding Acknowledgements
Type of funding sources: Private grant(s) and/or Sponsorship. Main funding source(s): British Heart Foundation
Introduction
Automated classification of arrhythmias in ECGs is becoming increasingly important. Publicly available ECG datasets have been widely used by the research community to create novel artificial intelligence models that improve these detection rates. The development of these models requires access to large volume of labelled data. However, access to such databases is becoming increasingly limited. In addition, the datasets are often unbalanced because abnormal rhythms are far outweighed by normal samples. The unbalanced nature of the datasets can lead to less accurate models. Therefore, generating realistic synthetic signals can augment the real signals found in such databases and provide data that allows sophisticated model development.
Purpose
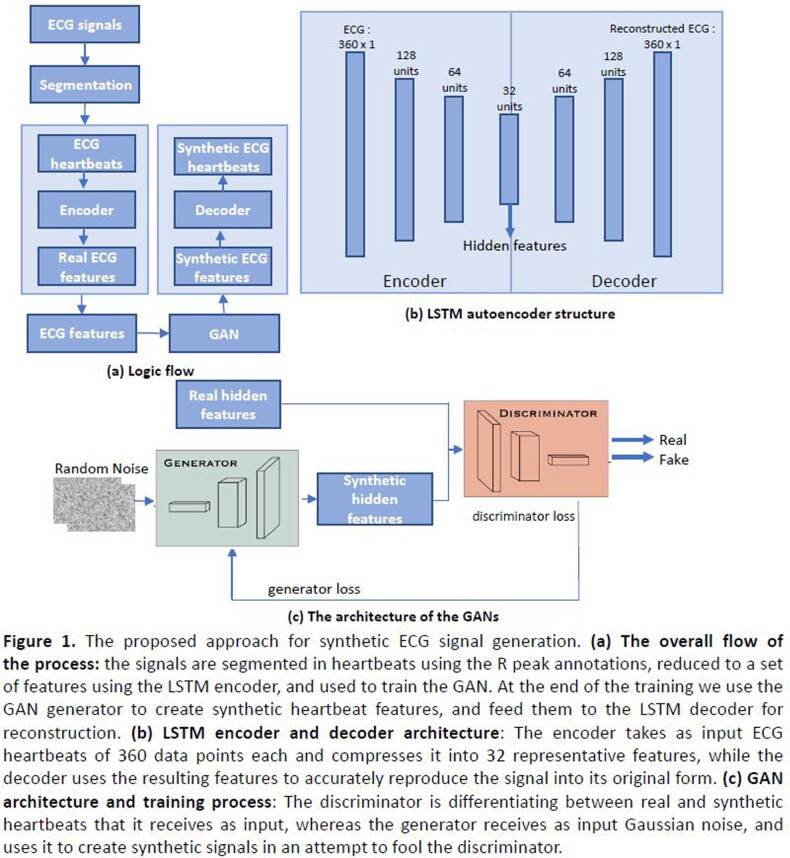
In this study, we propose a deep learning-based approach for synthetic ECG signal generation that uses long short-term memory (LSTM) autoencoder and generative adversarial networks (GAN) to generate signals that mimic the distribution of arrhythmia signals (Figure 1).
Methods
The LSTM autoencoder is composed of two parts: an encoder and a decoder (Figure 1b). The encoder takes original ECG signal as its input and uses LSTM layers to compress the signal into a set of features. The decoder is formed by reversing the encoding process, which uses the encoded features as its input and converts them back into the original signal.
To generate synthetic signals, we inserted GANs between the LSTM encoder and the decoder. GANs are composed of a generator and a discriminator (Figure 1c). The generator produces synthetic ECG features based on noise, whereas the discriminator tries to distinguish between real features and results received from the generator.
The pathological beats studies were: left bundle branch block (LBBB), right bundle branch block (RBBB), aberrated atrial premature (AA), and normal beats (N) from the MIT-BIH arrhythmia database, using lead II only.
To evaluate the quality of our synthetic signals, we trained an LSTM classifier on a combination of our real and synthetic data and compared the testing results with a model trained on real data alone.
Results
The LSTM encoder, decoder and GAN were trained individually for each beat type, and examples of generated signals are illustrated in Figure 2. The average accuracy of the classification for the original dataset was 90%, with a recall of 98% for N, 36% for AA, 39% for LBBB and 97% for RBBB. Once synthetic signals were added to the training set, the average testing classification accuracy increased to 98%, with a recall of 99% for N, 83% for AA, 99% for LBBB and 99% for RBBB.
For fair comparison, the testing set contained only real data and remained unchanged for both groups.