- Record: found
- Abstract: found
- Article: not found
A “Tug of War” Maintains a Dynamic Protein–Membrane Complex: Molecular Dynamics Simulations of C-Raf RBD-CRD Bound to K-Ras4B at an Anionic Membrane
Read this article at
Abstract

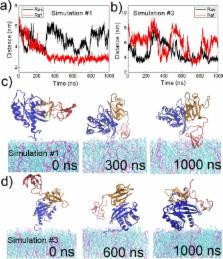
Association of Raf kinase with activated Ras triggers downstream signaling cascades toward regulating transcription in the cells’ nucleus. Dysregulation of Ras–Raf signaling stimulates cancers. We investigate the C-Raf RBD and CRD regions when bound to oncogenic K-Ras4B at the membrane. All-atom molecular dynamics simulations suggest that the membrane plays an integral role in regulating the configurational ensemble of the complex. Remarkably, the complex samples a few states dynamically, reflecting a competition between C-Raf CRD- and K-Ras4B- membrane interactions. This competition arises because the interaction between the RBD and K-Ras is strong while the linker between the RBD and CRD is short. Such a mechanism maintains a modest binding for the overall complex at the membrane and is expected to facilitate fast signaling processes. Competition of protein–membrane contacts is likely a common mechanism for other multiprotein complexes, if not multidomain proteins at membranes.
Abstract
As two counterparts of a protein complex, K-Ras4B and C-Raf CRD interact with membrane in a competitive manner due to topological restriction, presenting two major dynamic membrane-associated states.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
