- Record: found
- Abstract: found
- Article: found
Copy number variation of CCL3L1 among three major ethnic groups in Malaysia
Read this article at
Abstract
Background
C-C motif Chemokine Ligand 3 Like 1 ( CCL3L1) is a multiallelic copy number variable, which plays a crucial role in immunoregulatory and hosts defense through the production of macrophage inflammatory protein (MIP)-1α. Variable range of the CCL3L1 copies from 0 to 14 copies have been documented in several different populations. However, there is still lack of report on the range of CCL3L1 copy number exclusively among Malaysians who are a multi-ethnic population. Thus, this study aims to extensively examine the distribution of CCL3L1 copy number in the three major populations from Malaysia namely Malay, Chinese and Indian. A diploid copy number of CCL3L1 for 393 Malaysians (Malay = 178, Indian = 90, and Chinese = 125) was quantified using Paralogue Ratio Tests (PRTs) and then validated with microsatellites analysis.
Results
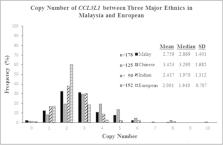
To our knowledge, this is the first report on the CCL3L1 copy number that has been attempted among Malaysians and the Chinese ethnic group exhibits a diverse pattern of CCL3L1 distribution copy number from the Malay and Indian ( p < 0.0001). The CCL3L1 ranged from 0 to 8 copies for both the Malay and Indian ethnic groups while 0 to 10 copies for the Chinese ethnic. Consequently, the CCL3L1 copy number among major ethnic groups in the Malaysian population is found to be significantly varied when compared to the European population ( p < 0.0001). The mean/median reported for the Malay, Chinese, Indian, and European are 2.759/2.869, 3.453/3.290, 2.437/1.970 and 2.001/1.940 respectively.
Conclusion
This study reveals the existence of genetic variation of CCL3L1 in the Malaysian population, and suggests by examining genetic diversity on the ethnicity, and specific geographical region could help in reconstructing human evolutionary history and for the prediction of disease risk related to the CCL3L1 copy number.
Related collections
Most cited references48
- Record: found
- Abstract: found
- Article: not found
The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility.
- Record: found
- Abstract: found
- Article: not found