- Record: found
- Abstract: found
- Article: found
Variants in the genes DCTN2, DNAH10, LRIG3, and MYO1A are associated with intermediate Charcot–Marie–Tooth disease in a Norwegian family
Read this article at
Abstract
Introduction
Charcot–Marie–Tooth disease ( CMT) is a heterogeneous inherited neuropathy. The number of known CMT genes is rapidly increasing mainly due to next‐generation sequencing technology, at present more than 70 CMT‐associated genes are known. We investigated whether variants in the DCTN2 could cause CMT.
Material and methods
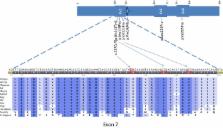
Fifty‐nine Norwegian CMT families from the general population with unknown genotype were tested by targeted next‐generation sequencing ( NGS) for variants in DCTN2 along with 32 CMT genes and 19 other genes causing other inherited neuropathies or neuronopathies, due to phenotypic overlap. In the family with the DCTN2 variant, exome sequencing was then carried out on all available eight family members to rule out the presence of more potential variants.
Results
Targeted NGS identified in one family a variant of DCTN2 , c.337C>T, segregating with the phenotype in five affected members, while it was not present in the three unaffected members. The DCTN2 variant c.337C>T; p.(His113Tyr) was neither found in in‐house controls nor in SNP databases. Exome sequencing revealed a singular heterozygous shared haplotype containing four genes, DCTN2 , DNAH10 , LRIG3, and MYO1A , with novel sequence variants. The haplotype was shared by all the affected members, while the unaffected members did not have it.
Related collections
Most cited references23
- Record: found
- Abstract: found
- Article: not found
Mutations in dynein link motor neuron degeneration to defects in retrograde transport.
- Record: found
- Abstract: found
- Article: not found
Charcot-Marie-Tooth disease subtypes and genetic testing strategies.
- Record: found
- Abstract: found
- Article: not found