- Record: found
- Abstract: found
- Article: found
Identification of a Novel Missense Mutation of the PHEX Gene in a Large Chinese Family with X-Linked Hypophosphataemia
Read this article at
Abstract
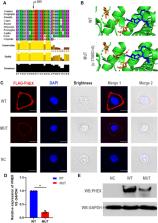
X-linked hypophosphataemia (XLH) is an X-linked dominant rare disease that refers to the most common hereditary hypophosphatemia (HH) caused by mutations in the phosphate-regulating endopeptidase homolog X-linked gene ( PHEX; OMIM: * 300550). However, mutations that have already been reported cannot account for all cases of XLH. Extensive genetic analysis can thus be helpful for arriving at the diagnosis of XLH. Herein, we identified a novel heterozygous mutation of PHEX (NM_000444.5: c.1768G > A) in a large Chinese family with XLH by whole-exome sequencing (WES). In addition, the negative effect of this mutation in PHEX was confirmed by both bioinformatics analysis and in vitro experimentation. The three-dimensional protein-model analysis predicted that this mutation might impair normal zinc binding. Immunofluorescence staining, qPCR, and western blotting analysis confirmed that the mutation we detected attenuated PHEX protein expression. The heterozygous mutation of PHEX (NM_000444.5: c.1768G > A) identified in this study by genetic and functional experiments constitutes a novel genetic cause of XLH, but further study will be required to expand its use in clinical and molecular diagnoses of XLH.
Related collections
Most cited references51

- Record: found
- Abstract: found
- Article: found
Highly accurate protein structure prediction with AlphaFold

- Record: found
- Abstract: found
- Article: found
AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models
- Record: found
- Abstract: found
- Article: not found