- Record: found
- Abstract: found
- Article: found
Promyelocytic leukemia protein (PML) controls breast cancer cell proliferation by modulating Forkhead transcription factors
Read this article at
Abstract
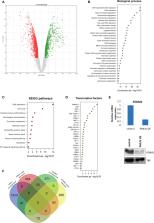
The multitasking promyelocytic leukemia ( PML) protein was originally recognized as a tumor‐suppressive factor, but more recent evidence has implicated PML in tumor cell prosurvival actions and poor patient prognosis in specific cancer settings. Here, we report that inducible PMLIV expression inhibits cell proliferation as well as self‐renewal and impairs cell cycle progression of breast cancer cell lines in a reversible manner. Transcriptomic profiling identified a large number of PML‐deregulated genes associated with various cell processes. Among them, cell cycle‐ and division‐related genes and their cognitive regulators are highly ranked. In this study, we focused on previously unknown PML targets, namely the Forkhead transcription factors. PML suppresses the Forkhead box subclass M1 ( FOXM1) transcription factor at both the RNA and protein levels, along with many of its gene targets. We show that FOXM1 interacts with PMLIV primarily via its DNA‐binding domain and dynamically colocalizes in PML nuclear bodies. In parallel, PML modulates the activity of Forkhead box O3 ( FOXO3), a factor opposing certain FOXM1 activities, to promote cell survival and stress resistance. Thus, PMLIV affects the balance of FOXO3 and FOXM1 transcriptional programs by acting on discrete gene subsets to favor both growth inhibition and survival. Interestingly, PMLIV‐specific knockdown mimicked ectopic expression vis‐à‐vis loss of proliferative ability and self‐renewal, but also led to loss of survival ability as shown by increased apoptosis. We propose that divergent or similar effects on cell physiology may be elicited by high or low PMLIV levels dictated by other concurrent genetic or epigenetic cancer cell states that may additionally account for its disparate effects in various cancer types.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
FoxM1 is required for execution of the mitotic programme and chromosome stability.
- Record: found
- Abstract: found
- Article: not found
Forkhead box proteins: tuning forks for transcriptional harmony.
- Record: found
- Abstract: found
- Article: not found