- Record: found
- Abstract: found
- Article: found
Genome-wide association study of delay discounting in Heterogenous Stock rats
Read this article at
Abstract
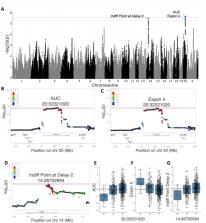
Delay discounting refers to the behavioral tendency to devalue rewards as a function of their delay in receipt. Heightened delay discounting has been associated with substance use disorders, as well as multiple co-occurring psychopathologies. Genetic studies in humans and animal models have established that delay discounting is a heritable trait, but only a few specific genes have been associated with delay discounting. Here, we aimed to identify novel genetic loci associated with delay discounting through a genome-wide association study (GWAS) using Heterogenous Stock rats, a genetically diverse outbred population derived from eight inbred founder strains. We assessed delay discounting in 650 male and female rats using an adjusting amount procedure in which rats chose between smaller immediate sucrose rewards or a larger reward at variable delays. Preference switch points were calculated for each rat and both exponential and hyperbolic functions were fitted to these indifference points. Area under the curve (AUC) and the discounting parameter k of both functions were used as delay discounting measures. GWAS for AUC, exponential k, and indifference points for a short delay identified significant loci on chromosomes 20 and 14. The gene Slc35f1, which encodes a member of the solute carrier family of nucleoside sugar transporters, was the only gene within the chromosome 20 locus. That locus also contained an eQTL for Slc35f1, suggesting that heritable differences in the expression of that gene might be responsible for the association with behavior. The gene Adgrl3, which encodes a member of the latrophilin family of G-protein coupled receptors, was the only gene within the chromosome 14 locus. These findings implicate novel genes in delay discounting and highlight the need for further exploration.
Related collections
Most cited references105

- Record: found
- Abstract: found
- Article: found
Fast and accurate short read alignment with Burrows–Wheeler transform
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads
- Record: found
- Abstract: found
- Article: not found