- Record: found
- Abstract: found
- Article: not found
Two Distinct Pathways Mediated by PA28 and hsp90 in Major Histocompatibility Complex Class I Antigen Processing
Read this article at
Abstract
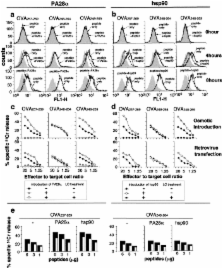
Major histocompatibility complex (MHC) class I ligands are mainly produced by the proteasome. Herein, we show that the processing of antigens is regulated by two distinct pathways, one requiring PA28 and the other hsp90. Both hsp90 and PA28 enhanced the antigen processing of ovalbumin (OVA). Geldanamycin, an inhibitor of hsp90, almost completely suppressed OVA antigen presentation in PA28α −/−/β −/− lipopolysaccharide blasts, but not in wild-type cells, indicating that hsp90 compensates for the loss of PA28 and is essential in the PA28-independent pathway. In contrast, treatment of cells with interferon (IFN)-γ, which induces PA28 expression, abrogated the requirement of hsp90, suggesting that IFN-γ enhances the PA28-dependent pathway, whereas it diminishes hsp90-dependent pathway. Importantly, IFN-γ did not induce MHC class I expressions in PA28-deficient cells, indicating a prominent role for PA28 in IFN-γ–stimulated peptide supply. Thus, these two pathways operate either redundantly or specifically, depending on antigen species and cell type.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
The 26S proteasome: a molecular machine designed for controlled proteolysis.
- Record: found
- Abstract: found
- Article: not found
Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone.
- Record: found
- Abstract: found
- Article: not found
