- Record: found
- Abstract: found
- Article: found
U2AF1 and EZH2 mutations are associated with nonimmune hemolytic anemia in myelodysplastic syndromes

Read this article at
Key Points
Visual Abstract
Abstract
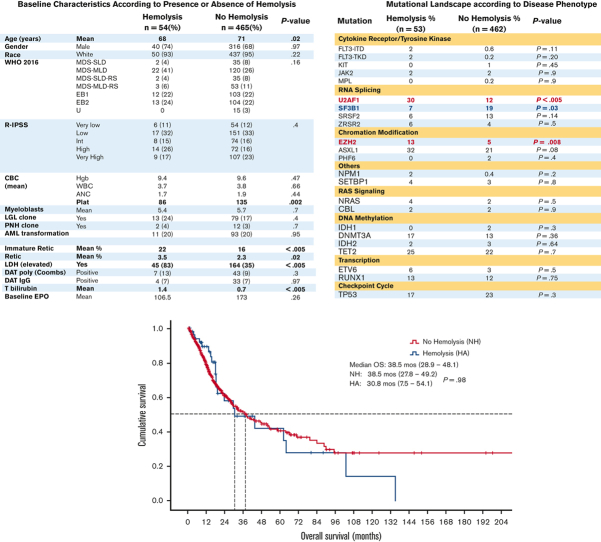
Hemolysis is a well-recognized but poorly characterized phenomenon in a subset of patients with myelodysplastic syndromes (MDS). Its pathobiological basis seems to underpin a nonimmune etiology whose clinical significance has not been adequately characterized. Hemolysis in MDS is often attributed to either ineffective intramedullary erythropoiesis or acquired hemoglobinopathies and red blood cell (RBC) membrane defects. These heterogeneous processes have not been associated with specific genetic subsets of the disease. We aimed to describe the prevalence of hemolysis among patients with MDS, their baseline characteristics, molecular features, and resulting impact on outcomes. We considered baseline serum haptoglobin <10 mg/dL a surrogate marker for intravascular hemolysis. Among 519 patients, 10% had hemolysis. The baseline characteristics were similar among both groups. Only 13% of patients with hemolysis were Coombs-positive, suggesting that hemolysis in MDS is largely not immune-mediated. Inferior survival trends were observed among lower-risk patients with MDS undergoing hemolysis. Decreased response rates to erythropoiesis-stimulating agents (ESA) and higher responses to hypomethylating agents (HMA) were also observed in the hemolysis group. U2AF1 and EZH2 hotspot mutations were more prevalent among those undergoing hemolysis ( P < .05). U2AF1 mutations were observed in 30% of patients with hemolysis and occurred almost exclusively at the S34 hotspot. Somatic mutations encoding splicing factors may affect erythrocyte membrane components, biochemical properties, and RBC metabolic function, which underpin the development of atypical clones from erythroid precursors in MDS presenting with hemolysis. Future studies will explore the contribution of altered splicing to the development of acquired hemoglobinopathies.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Clinical effect of point mutations in myelodysplastic syndromes.
- Record: found
- Abstract: found
- Article: not found
Clinical and biological implications of driver mutations in myelodysplastic syndromes.
- Record: found
- Abstract: found
- Article: found