- Record: found
- Abstract: found
- Article: found
Early stress-induced impaired microglial pruning of excitatory synapses on immature CRH-expressing neurons provokes aberrant adult stress responses
Read this article at
SUMMARY
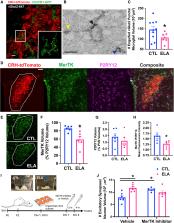
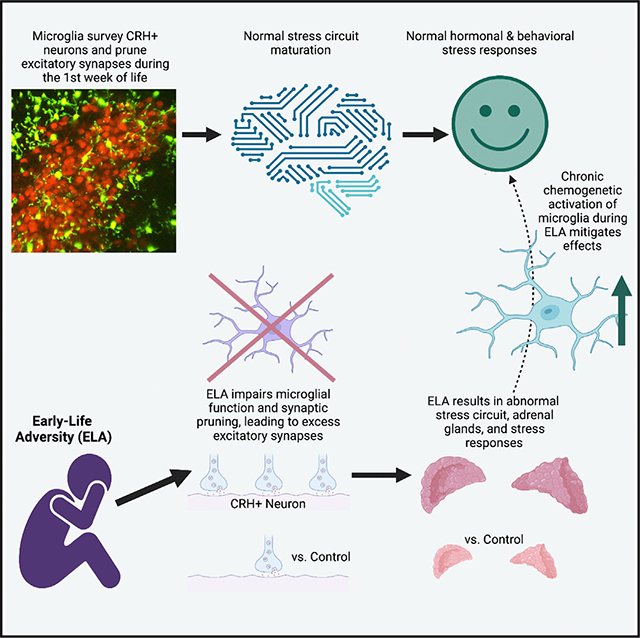
Several mental illnesses, characterized by aberrant stress reactivity, often arise after early-life adversity (ELA). However, it is unclear how ELA affects stress-related brain circuit maturation, provoking these enduring vulnerabilities. We find that ELA increases functional excitatory synapses onto stress-sensitive hypothalamic corticotropin-releasing hormone (CRH)-expressing neurons, resulting from disrupted developmental synapse pruning by adjacent microglia. Microglial process dynamics and synaptic element engulfment were attenuated in ELA mice, associated with deficient signaling of the microglial phagocytic receptor MerTK. Accordingly, selective chronic chemogenetic activation of ELA microglia increased microglial process dynamics and reduced excitatory synapse density to control levels. Notably, selective early-life activation of ELA microglia normalized adult acute and chronic stress responses, including stress-induced hormone secretion and behavioral threat responses, as well as chronic adrenal hypertrophy of ELA mice. Thus, microglial actions during development are powerful contributors to mechanisms by which ELA sculpts the connectivity of stress-regulating neurons, promoting vulnerability to stress and stress-related mental illnesses.
In brief
Early-life adversity (ELA) promotes lifelong aberrant stress responses and vulnerability to mental illnesses. Bolton et al. identify poor dynamics and hypothalamic CRH neurons’ excitatory synapse pruning of ELA microglia, implicating microglial MerTK. Chronic chemogenetic activation of ELA microglia normalized process dynamics, synapse density, and adult hormonal and behavioral stress responses.
Graphical Abstract
Related collections
Most cited references112
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.

- Record: found
- Abstract: found
- Article: found
SciPy 1.0: fundamental algorithms for scientific computing in Python
- Record: found
- Abstract: found
- Article: not found