- Record: found
- Abstract: found
- Article: not found
Cell-Nonautonomous Regulation of C. elegans Germ Cell Death by kri-1
Read this article at
Summary
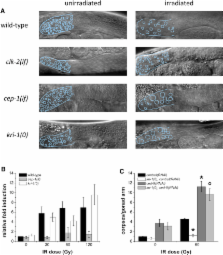
Programmed cell death (or apoptosis) is an evolutionarily conserved, genetically controlled suicide mechanism for cells that, when deregulated, can lead to developmental defects, cancers, and degenerative diseases [1, 2]. In C. elegans, DNA damage induces germ cell death by signaling through cep-1/ p53, ultimately leading to the activation of CED-3/caspase [3–13]. It has been hypothesized that the major regulatory events controlling cell death occur by cell-autonomous mechanisms, that is, within the dying cell. In support of this, genetic studies in C. elegans have shown that the core apoptosis pathway genes ced-4/ APAF-1 and ced-3/caspase are required in cells fated to die [9]. However, it is not known whether the upstream signals that activate apoptosis function in a cell-autonomous manner. Here we show that kri-1, an ortholog of KRIT1/ CCM1, which is mutated in the human neurovascular disease cerebral cavernous malformation [14, 15], is required to activate DNA damage-dependent cell death independently of cep-1/ p53. Interestingly, we find that kri-1 regulates cell death in a cell-nonautonomous manner, revealing a novel regulatory role for nondying cells in eliciting cell death in response to DNA damage.
Highlights
► kri-1 is a novel positive regulator of C. elegans germ cell apoptosis ► kri-1 regulates cell death independently of cep-1/ p53 ► kri-1 regulates cell death cell nonautonomously, possibly by cross-tissue signaling
Related collections
Most cited references28
- Record: found
- Abstract: found
- Article: not found
On the role of RNA amplification in dsRNA-triggered gene silencing.
- Record: found
- Abstract: found
- Article: not found
A conserved checkpoint pathway mediates DNA damage--induced apoptosis and cell cycle arrest in C. elegans.
- Record: found
- Abstract: found
- Article: not found
