- Record: found
- Abstract: found
- Article: found
A Novel Precision Approach to Overcome the “Addiction Pandemic” by Incorporating Genetic Addiction Risk Severity (GARS) and Dopamine Homeostasis Restoration
Read this article at
Abstract
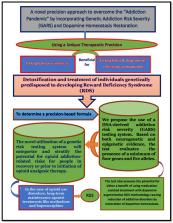
This article describes a unique therapeutic precision intervention, a formulation of enkephalinase inhibitors, enkephalin, and dopamine-releasing neuronutrients, to induce dopamine homeostasis for detoxification and treatment of individuals genetically predisposed to developing reward deficiency syndrome (RDS). The formulations are based on the results of the addiction risk severity (GARS) test. Based on both neurogenetic and epigenetic evidence, the test evaluates the presence of reward genes and risk alleles. Existing evidence demonstrates that the novel genetic risk testing system can successfully stratify the potential for developing opioid use disorder (OUD) related risks or before initiating opioid analgesic therapy and RDS risk for people in recovery. In the case of opioid use disorders, long-term maintenance agonist treatments like methadone and buprenorphine may create RDS, or RDS may have been in existence, but not recognized. The test will also assess the potential for benefit from medication-assisted treatment with dopamine augmentation. RDS methodology holds a strong promise for reducing the burden of addictive disorders for individuals, their families, and society as a whole by guiding the restoration of dopamine homeostasisthrough anti-reward allostatic neuroadaptations. WC 175.
Related collections
Most cited references112
- Record: found
- Abstract: found
- Article: found
Identification of common genetic risk variants for autism spectrum disorder
- Record: found
- Abstract: found
- Article: not found
A global overview of pleiotropy and genetic architecture in complex traits
- Record: found
- Abstract: found
- Article: not found