- Record: found
- Abstract: found
- Article: found
Eukaryotic CD-NTase, STING, and viperin proteins evolved via domain shuffling, horizontal transfer, and ancient inheritance from prokaryotes
Read this article at
Abstract
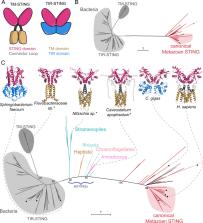
Animals use a variety of cell-autonomous innate immune proteins to detect viral infections and prevent replication. Recent studies have discovered that a subset of mammalian antiviral proteins have homology to antiphage defense proteins in bacteria, implying that there are aspects of innate immunity that are shared across the Tree of Life. While the majority of these studies have focused on characterizing the diversity and biochemical functions of the bacterial proteins, the evolutionary relationships between animal and bacterial proteins are less clear. This ambiguity is partly due to the long evolutionary distances separating animal and bacterial proteins, which obscures their relationships. Here, we tackle this problem for 3 innate immune families (CD-NTases [including cGAS], STINGs, and viperins) by deeply sampling protein diversity across eukaryotes. We find that viperins and OAS family CD-NTases are ancient immune proteins, likely inherited since the earliest eukaryotes first arose. In contrast, we find other immune proteins that were acquired via at least 4 independent events of horizontal gene transfer (HGT) from bacteria. Two of these events allowed algae to acquire new bacterial viperins, while 2 more HGT events gave rise to distinct superfamilies of eukaryotic CD-NTases: the cGLR superfamily (containing cGAS) that has since diversified via a series of animal-specific duplications and a previously undefined eSMODS superfamily, which more closely resembles bacterial CD-NTases. Finally, we found that cGAS and STING proteins have substantially different histories, with STING protein domains undergoing convergent domain shuffling in bacteria and eukaryotes. Overall, our findings paint a picture of eukaryotic innate immunity as highly dynamic, where eukaryotes build upon their ancient antiviral repertoires through the reuse of protein domains and by repeatedly sampling a rich reservoir of bacterial antiphage genes.
Abstract
How and when did our innate immune systems first evolve? This study analyses diverse eukaryotes, uncovering the evolutionary origins of three families of antiviral proteins: CD-NTases (including cGAS), STINGs and Viperins; each reveals a different story connecting animal and bacterial immunity.
Related collections
Most cited references95

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
Highly accurate protein structure prediction with AlphaFold
- Record: found
- Abstract: found
- Article: not found