- Record: found
- Abstract: found
- Article: found
Mikania micrantha genome provides insights into the molecular mechanism of rapid growth
Read this article at
Abstract
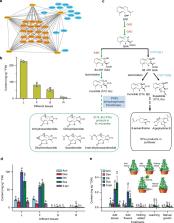
Mikania micrantha is one of the top 100 worst invasive species that can cause serious damage to natural ecosystems and substantial economic losses. Here, we present its 1.79 Gb chromosome-scale reference genome. Half of the genome is composed of long terminal repeat retrotransposons, 80% of which have been derived from a significant expansion in the past one million years. We identify a whole genome duplication event and recent segmental duplications, which may be responsible for its rapid environmental adaptation. Additionally, we show that M. micrantha achieves higher photosynthetic capacity by CO 2 absorption at night to supplement the carbon fixation during the day, as well as enhanced stem photosynthesis efficiency. Furthermore, the metabolites of M. micrantha can increase the availability of nitrogen by enriching the microbes that participate in nitrogen cycling pathways. These findings collectively provide insights into the rapid growth and invasive adaptation.
Abstract
Mikania micrantha is an extremely fast-growing invasive plant species that can cause serious damage to natural ecosystems. Here, the authors assemble its chromosome-scale reference genome and explore possible mechanisms that contribute to its rapid growth.
Related collections
Most cited references47

- Record: found
- Abstract: found
- Article: found
KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies
- Record: found
- Abstract: found
- Article: not found
Synteny and collinearity in plant genomes.
- Record: found
- Abstract: found
- Article: not found