- Record: found
- Abstract: found
- Article: found
Mechanistic convergence of the TIGIT and PD-1 inhibitory pathways necessitates co-blockade to optimize anti-tumor CD8 + T cell responses

Read this article at
SUMMARY
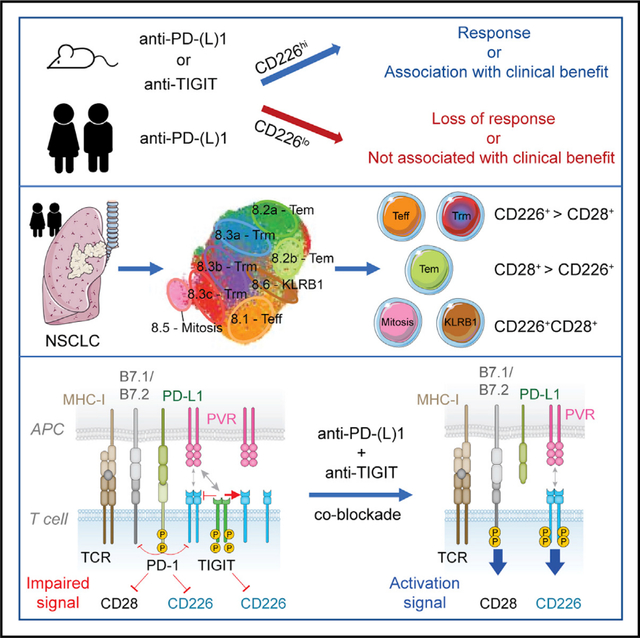
Dual blockade of the PD-1 and TIGIT coinhibitory receptors on T cells shows promising early results in cancer patients. Here, we studied the mechanisms whereby PD-1 and/or TIGIT blockade modulate anti-tumor CD8 + T cells. Although PD-1 and TIGIT are thought to regulate different costimulatory receptors (CD28 and CD226), effectiveness of PD-1 or TIGIT inhibition in preclinical tumor models was reduced in the absence of CD226. CD226 expression associated with clinical benefit in patients with non-small cell lung carcinoma (NSCLC) treated with anti-PD-L1 antibody atezolizumab. CD226 and CD28 were co-expressed on NSCLC infiltrating CD8 + T cells poised for expansion. Mechanistically, PD-1 inhibited phosphorylation of both CD226 and CD28 via its ITIM-containing intracellular domain (ICD); TIGIT’s ICD was dispensable, with TIGIT restricting CD226 co-stimulation by blocking interaction with their common ligand PVR (CD155). Thus, full restoration of CD226 signaling, and optimal anti-tumor CD8 + T cell responses, requires blockade of TIGIT and PD-1, providing a mechanistic rationale for combinatorial targeting in the clinic.
Graphical Abstract
In brief
Dual blockade of the PD-1 and TIGIT coinhibitory receptors on T cells shows promising early results in cancer patients. Banta et al. find that PD-1 and TIGIT disrupt activation of the costimulatory receptor CD226 through distinct mechanisms, providing mechanistic rationale for the dual blockade of PD-(L)1 and TIGIT in cancer immunotherapy.
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data
- Record: found
- Abstract: found
- Article: not found