- Record: found
- Abstract: found
- Article: found
Effect of lactulose intervention on gut microbiota and short chain fatty acid composition of C57BL/6J mice
Read this article at
Abstract
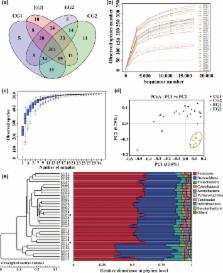
Gut microbiota have strong connections with health. Lactulose has been shown to regulate gut microbiota and benefit host health. In this study, the effect of short‐term (3 week) intervention of lactulose on gut microbiota was investigated. Gut microbiota were detected from mouse feces by 16S rRNA high‐throughput sequencing, and short chain fatty acids ( SCFAs) were detected by gas chromatography‐mass spectrometry ( GC‐ MS). Lactulose intervention enhanced the α‐diversity of the gut microbiota; increased the abundance of hydrogen‐producing bacteria Prevotellaceae and Rikenellaceae, probiotics Bifidobacteriaceae and Lactobacillaceae, and mucin‐degrading bacteria Akkermansia and Helicobacter; decreased the abundance of harmful bacteria Desulfovibrionaceae and branched‐chain SCFAs ( BCFAs). These results suggest that lactulose intervention effectively increased the diversity and improved the structure of the intestinal microbiota, which may be beneficial for host health.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
The influence of diet on the gut microbiota.

- Record: found
- Abstract: found
- Article: found
Towards microbial fermentation metabolites as markers for health benefits of prebiotics

- Record: found
- Abstract: found
- Article: found
A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.