- Record: found
- Abstract: found
- Article: found
An optogenetic toolbox of LOV-based photosensitizers for light-driven killing of bacteria
Read this article at
Abstract
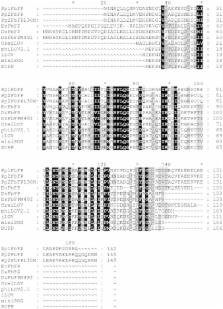
Flavin-binding fluorescent proteins (FPs) are genetically encoded in vivo reporters, which are derived from microbial and plant LOV photoreceptors. In this study, we comparatively analyzed ROS formation and light-driven antimicrobial efficacy of eleven LOV-based FPs. In particular, we determined singlet oxygen ( 1O 2) quantum yields and superoxide photosensitization activities via spectroscopic assays and performed cell toxicity experiments in E. coli. Besides miniSOG and SOPP, which have been engineered to generate 1O 2, all of the other tested flavoproteins were able to produce singlet oxygen and/or hydrogen peroxide but exhibited remarkable differences in ROS selectivity and yield. Accordingly, most LOV-FPs are potent photosensitizers, which can be used for light-controlled killing of bacteria. Furthermore, the two variants Pp2FbFP and DsFbFP M49I, exhibiting preferential photosensitization of singlet oxygen or singlet oxygen and superoxide, respectively, were shown to be new tools for studying specific ROS-induced cell signaling processes. The tested LOV-FPs thus further expand the toolbox of optogenetic sensitizers usable for a broad spectrum of microbiological and biomedical applications.
Related collections
Most cited references91
- Record: found
- Abstract: found
- Article: not found
Photodynamic therapy (PDT): a short review on cellular mechanisms and cancer research applications for PDT.
- Record: found
- Abstract: found
- Article: not found
Bright monomeric red fluorescent protein with an extended fluorescence lifetime.
- Record: found
- Abstract: found
- Article: not found