- Record: found
- Abstract: found
- Article: found
The DNMT3A R882H mutation does not cause dominant negative effects in purified mixed DNMT3A/R882H complexes
Read this article at
Abstract
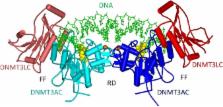
The DNA methyltransferase DNMT3A R882H mutation is observed in 25% of all AML patients. DNMT3A is active as tetramer and the R882H mutation is located in one of the subunit/subunit interfaces. Previous work has reported that formation of mixed wildtype/R882H complexes leads to a strong loss of catalytic activity observed in in vitro DNA methylation assays (Russler-Germain et al., 2014, Cancer Cell 25:442–454). To investigate this effect further, we have prepared mixed wildtype/R882H DNMT3A complexes by incubation of individually purified subunits of the DNMT3A catalytic domain and full-length DNMT3A2. In addition, we have used a double affinity tag approach and specifically purified mixed catalytic domain complexes formed after co-expression of R882H and wildtype subunits in E. coli cells. Afterwards, we determined the catalytic activity of the mixed complexes and compared it to that of purified complexes only consisting of one subunit type. In both settings, the expected catalytic activities of mixed R882H/wildtype complexes were observed demonstrating an absence of a dominant negative effect of the R882H mutation in purified DNMT3A enzymes. This result suggests that heterocomplex formation of DNMT3A and R882H is unlikely to cause dominant negative effects in human cells as well. The limitations of this conclusion and its implications are discussed.
Related collections
Most cited references28

- Record: found
- Abstract: found
- Article: found
Role of TET enzymes in DNA methylation, development, and cancer
- Record: found
- Abstract: found
- Article: not found
Cancer genetics and epigenetics: two sides of the same coin?
- Record: found
- Abstract: found
- Article: not found