- Record: found
- Abstract: found
- Article: found
Putative Novel Effector Genes Revealed by the Genomic Analysis of the Phytopathogenic Fungus Fusarium oxysporum f. sp. physali ( Foph) That Infects Cape Gooseberry Plants
Read this article at
Abstract
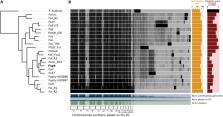
The vascular wilt disease caused by the fungus Fusarium oxysporum f. sp. physali ( Foph) is one of the most limiting factors for the production and export of cape gooseberry ( Physalis peruviana) in Colombia. A transcriptomic analysis of a highly virulent strain of F. oxysporum in cape gooseberry plants, revealed the presence of secreted in the xylem (SIX) effector genes, known to be involved in the pathogenicity of other formae speciales (ff. spp.) of F. oxysporum. This pathogenic strain was classified as a new f. sp. named Foph, due to its specificity for cape gooseberry hosts. Here, we sequenced and assembled the genome of five strains of F. oxysporum from a fungal collection associated to the cape gooseberry crop (including Foph), focusing on the validation of the presence of SIX homologous and on the identification of putative effectors unique to Foph. By comparative and phylogenomic analyses based on single-copy orthologous, we found that Foph is closely related to F. oxysporum ff. spp., associated with solanaceous hosts. We confirmed the presence of highly identical homologous genomic regions between Foph and Fol that contain effector genes and identified six new putative effector genes, specific to Foph pathogenic strains. We also conducted a molecular characterization using this set of putative novel effectors in a panel of 36 additional stains of F. oxysporum including two of the four sequenced strains, from the fungal collection mentioned above. These results suggest the polyphyletic origin of Foph and the putative independent acquisition of new candidate effectors in different clades of related strains. The novel effector candidates identified in this genomic analysis, represent new sources involved in the interaction between Foph and cape gooseberry, that could be implemented to develop appropriate management strategies of the wilt disease caused by Foph in the cape gooseberry crop.
Related collections
Most cited references72

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
Basic local alignment search tool.
- Record: found
- Abstract: found
- Article: not found