- Record: found
- Abstract: found
- Article: found
From Molecular Descriptors to Intrinsic Fish Toxicity of Chemicals: An Alternative Approach to Chemical Prioritization
Read this article at
Abstract
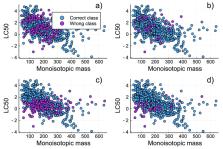
The European and U.S. chemical agencies have listed approximately 800k chemicals about which knowledge of potential risks to human health and the environment is lacking. Filling these data gaps experimentally is impossible, so in silico approaches and prediction are essential. Many existing models are however limited by assumptions (e.g., linearity and continuity) and small training sets. In this study, we present a supervised direct classification model that connects molecular descriptors to toxicity. Categories can be driven by either data (using k-means clustering) or defined by regulation. This was tested via 907 experimentally defined 96 h LC 50 values for acute fish toxicity. Our classification model explained ≈90% of the variance in our data for the training set and ≈80% for the test set. This strategy gave a 5-fold decrease in the frequency of incorrect categorization compared to a quantitative structure–activity relationship (QSAR) regression model. Our model was subsequently employed to predict the toxicity categories of ≈32k chemicals. A comparison between the model-based applicability domain (AD) and the training set AD was performed, suggesting that the training set-based AD is a more adequate way to avoid extrapolation when using such models. The better performance of our direct classification model compared to that of QSAR methods makes this approach a viable tool for assessing the hazards and risks of chemicals.
Abstract
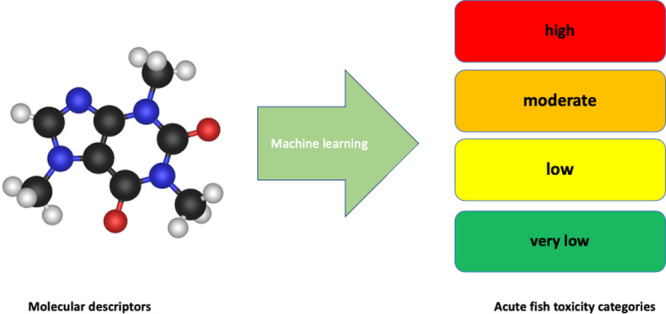
In this study, a machine learning-based strategy that is an alternative to conventional quantitative structure−activity relationship models is used for the toxicity categorization of chemicals using molecular descriptors and direct classification.
Related collections
Most cited references49
- Record: found
- Abstract: not found
- Article: not found
SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules
- Record: found
- Abstract: found
- Article: not found
Extended-connectivity fingerprints.
- Record: found
- Abstract: found
- Article: not found