- Record: found
- Abstract: found
- Article: found
Germline Mutation in MUS81 Resulting in Impaired Protein Stability is Associated with Familial Breast and Thyroid Cancer
Read this article at
Abstract
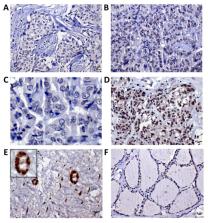
Multiple primary thyroid cancer (TC) and breast cancer (BC) are commonly diagnosed, and the lifetime risk for these cancers is increased in patients with a positive family history of both TC and BC. Although this phenotype is partially explained by TP53 or PTEN mutations, a significant number of patients are negative for these alterations. We judiciously recruited patients diagnosed with BC and/or TC having a family history of these tumors and assessed their whole-exome sequencing. After variant prioritization, we selected MUS81 c.1292G>A (p.R431H) for further investigation. This variant was genotyped in a healthy population and sporadic BC/TC tissues and investigated at the protein level and cellular models. MUS81 c.1292G>A was the most frequent variant (25%) and the strongest candidate due to its function of double-strand break repair. This variant was confirmed in four relatives from two families. MUS81 p.R431H protein exhibited lower expression levels in tumors from patients positive for the germline variant, compared with wild-type BC, and normal breast and thyroid tissues. Using cell line models, we showed that c.1292G>A induced protein instability and affected DNA damage response. We suggest that MUS81 is a novel candidate involved in familial BC/TC based on its low frequency in healthy individuals and proven effect in protein stability.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
Realizing the promise of cancer predisposition genes.

- Record: found
- Abstract: found
- Article: found
In silico prediction of splice-altering single nucleotide variants in the human genome
- Record: found
- Abstract: found
- Article: not found