- Record: found
- Abstract: found
- Article: found
A Quantum Chemical and Statistical Study of Cytotoxic Activity of Compounds Isolated from Curcuma zedoaria
Read this article at
Abstract
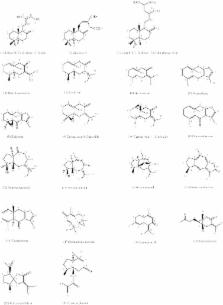
A series of 21 compounds isolated from Curcuma zedoaria was subjected to cytotoxicity test against MCF7; Ca Ski; PC3 and HT-29 cancer cell lines; and a normal HUVEC cell line. To rationalize the structure–activity relationships of the isolated compounds; a set of electronic; steric and hydrophobic descriptors were calculated using density functional theory (DFT) method. Statistical analyses were carried out using simple and multiple linear regressions (SLR; MLR); principal component analysis (PCA); and hierarchical cluster analysis (HCA). SLR analyses showed that the cytotoxicity of the isolated compounds against a given cell line depend on certain descriptors; and the corresponding correlation coefficients (R 2) vary from 0%–55%. MLR results revealed that the best models can be achieved with a limited number of specific descriptors applicable for compounds having a similar basic skeleton. Based on PCA; HCA and MLR analyses; active compounds were classified into subgroups; which was in agreement with the cell based cytotoxicity assay.
Related collections
Most cited references43
- Record: found
- Abstract: not found
- Article: not found
A new mixing of Hartree–Fock and local density-functional theories
- Record: found
- Abstract: not found
- Article: not found
Applied multivariate statistical analysis
- Record: found
- Abstract: found
- Article: not found