- Record: found
- Abstract: found
- Article: found
Cytometry of chromatin bound Mcm6 and PCNA identifies two states in G1 that are separated functionally by the G1 restriction point 1
Read this article at
Abstract
Background
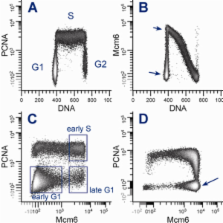
Cytometric measurements of DNA content and chromatin-bound Mcm2 have demonstrated bimodal patterns of expression in G1. These patterns, the replication licensing function of Mcm proteins, and a correlation between Mcm loading and cell cycle commitment for cells re-entering the cell cycle, led us to test the idea that cells expressing a defined high level of chromatin-bound Mcm6 in G1 are committed - i.e., past the G1 restriction point. We developed a cell-based assay for tightly-bound PCNA (PCNA*) and Mcm6 (Mcm6*), DNA content, and a mitotic marker to clearly define G1, S, G2, and M phases of the cell cycle. hTERT-BJ1, hTERT-RPE-1, and Molt4 cells were extracted with Triton X-100 followed by methanol fixation, stained with antibodies and DAPI, then measured by cytometry.
Results
Bivariate analysis of cytometric data demonstrated complex patterns with distinct clustering for all combinations of the 4 variables. In G1, cells clustered in two groups characterized by low and high Mcm6* expression. Serum starvation and release experiments showed that residence in the high group was in late G1, just prior to S phase. Kinetic experiments, employing serum withdrawal, and stathmokinetic analysis with aphidicolin, mimosine or nocodazole demonstrated that cells with high levels of Mcm6* cycled with the committed phases of the cell cycle (S, G2, and M).
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication.
- Record: found
- Abstract: found
- Article: not found
The Pezcoller lecture: cancer cell cycles revisited.
- Record: found
- Abstract: found
- Article: not found