- Record: found
- Abstract: found
- Article: found
Expanding the Knowledge on the Skillful Yeast Cyberlindnera jadinii
Read this article at
Abstract
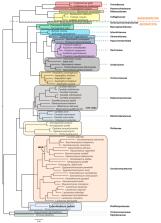
Cyberlindnera jadinii is widely used as a source of single-cell protein and is known for its ability to synthesize a great variety of valuable compounds for the food and pharmaceutical industries. Its capacity to produce compounds such as food additives, supplements, and organic acids, among other fine chemicals, has turned it into an attractive microorganism in the biotechnology field. In this review, we performed a robust phylogenetic analysis using the core proteome of C. jadinii and other fungal species, from Asco- to Basidiomycota, to elucidate the evolutionary roots of this species. In addition, we report the evolution of this species nomenclature over-time and the existence of a teleomorph ( C. jadinii) and anamorph state ( Candida utilis) and summarize the current nomenclature of most common strains. Finally, we highlight relevant traits of its physiology, the solute membrane transporters so far characterized, as well as the molecular tools currently available for its genomic manipulation. The emerging applications of this yeast reinforce its potential in the white biotechnology sector. Nonetheless, it is necessary to expand the knowledge on its metabolism, regulatory networks, and transport mechanisms, as well as to develop more robust genetic manipulation systems and synthetic biology tools to promote the full exploitation of C. jadinii.
Related collections
Most cited references144
- Record: found
- Abstract: found
- Article: not found
CRISPR-Cas systems for editing, regulating and targeting genomes.
- Record: found
- Abstract: not found
- Article: not found
Life with 6000 Genes
- Record: found
- Abstract: found
- Article: not found