- Record: found
- Abstract: found
- Article: found
Hypermethylation of the CTRP9 promoter region promotes Hcy induced VSMC lipid deposition and foam cell formation via negatively regulating ER stress
Read this article at
Abstract
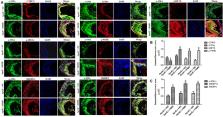
To provide a theoretical basis for the prevention and treatment of atherosclerosis (As), the current study aimed to investigate the mechanism underlying the effect of homocysteine (Hcy) on inducing the lipid deposition and foam cell formation of the vascular smooth muscle cell (VSMC) via C1q/Tumor necrosis factor-related protein9 (CTRP9) promoter region Hypermethylation negative regulating endoplasmic reticulum stress (ERs). Therefore, apolipoprotein E deficient (ApoE −/−) mice were randomly divided into the control [ApoE −/− + normal diet (NC)] and high methionine [ApoE −/− + (normal diet supplemented with 1.7% methionine (HMD)] groups (n = 6 mice/group). Following feeding for 15 weeks, the serum levels of Homocysteine (Hcy), total cholesterol (TC), and triglyceride (TG) were measured using an automatic biochemical analyzer. HE and oil red O staining were performed on the aorta roots to observe the pathological changes. Additionally, immunofluorescence staining was performed to detect the protein expression levels of CTRP9, glucose-regulated protein 78 kD (GRP78), phosphorylated protein kinase RNA-like ER kinase (p-PERK), activating transcription factor 6a (ATF6a), phosphorylated inositol-requiring enzyme-1α (p-IRE1α), sterol regulatory element binding proteins-1c (SREBP1c) and sterol regulatory element binding proteins-2 (SREBP2) in VSMC derived from murine aortic roots. In vitro, VSMC was stimulated with 100 μmol/l Hcy. After transfection of plasmids with overexpression and interference of CTRP9, ERs agonist (TM) and inhibitor (4-PBA) were given to stimulate VSMC cells. HE staining and oil red O staining were used to observe the effect of Hcy stimulation on lipid deposition in VSMC. Additionally, The mRNA and protein expression levels of CTRP9, GRP78, PERK, ATF6a, IRE1α, SREBP1c, and SREBP2 in VSMC were detected by RT-qPCR and western blot analysis, respectively. Finally, The methylation modification of the CTRP9 promoter region has been studied. The NCBI database was used to search the promoter region of the CTRP9 gene, and CpG Island was used to predict the methylation site. After Hcy stimulation of VSMC, overexpression of DNMT1, and intervention with 5-Azc, assess the methylation level of the CTRP9 promoter through bisulfite sequencing PCR (BSP). The results showed that the serum levels of Hcy, TC, and TG in the ApoE −/− + HMD group were significantly increased compared with the ApoE −/− + NC group. In addition, HE staining and oil red O staining showed obvious AS plaque formation in the vessel wall, and a large amount of fat deposition in VSMC, thus indicating that the hyperhomocysteinemia As an animal model was successfully established. Furthermore, CTRP9 were downregulated, while GRP78, p-PERK, ATF6a, p-IRE1α, SREBP1c, SREBP2 was upregulated in aortic VSMC in the ApoE −/− + HMD group. Consistent with the in vivo results, Hcy can inhibit the expression of CTRP9 in VSMC and induce ERs and lipid deposition in VSMC. Meanwhile, the increased expression of CTRP9 can reduce ERs and protect the lipid deposition in Hcy induced VSMC. Furthermore, ERs can promote Hcy induced VSMC lipid deposition, inhibition of ERs can reduce Hcy induced VSMC lipid deposition, and CTRP9 may play a protective role in Hcy induced VSMC lipid deposition and foam cell transformation through negative regulation of ERs. In addition, The CTRP9 promoter in the Hcy group showed hypermethylation. At the same time as Hcy intervention, overexpression of DNMT1 increases the methylation level of the CTRP9 promoter, while 5-Azc can reduce the methylation level of the CTRP9 promoter. Finally, Hcy can up-regulate the expression of DNMT1 and down-regulate the expression of CTRP9. After overexpression of DNMT1, the expression of CTRP9 is further decreased. After 5-Azc inhibition of DNMT1, the expression of DNMT1 decreases, while the expression of CTRP9 increases. It is suggested that the molecular mechanism of Hcy inhibiting the expression of CTRP9 is related to the hypermethylation of the CTRP9 promoter induced by Hcy and regulated by DNMT1. 5-Azc can inhibit the expression of DNMT1 and reverse the regulatory effect of DNMT1 on CTRP9. Overall, the results of the present study suggested that Hcy induces DNA hypermethylation in the CTRP9 promoter region by up-regulating DNMT1 expression, and negatively regulates ERs mediated VSMC lipid deposition and foam cell formation. CTRP9 may potentially be a therapeutic target in the treatment of hyperhomocysteinemia and As.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
DNA methylation and its basic function.
- Record: found
- Abstract: found
- Article: not found