- Record: found
- Abstract: found
- Article: found
Metabolomics for personalized medicine: the input of analytical chemistry from biomarker discovery to point-of-care tests
Read this article at
Graphical abstract
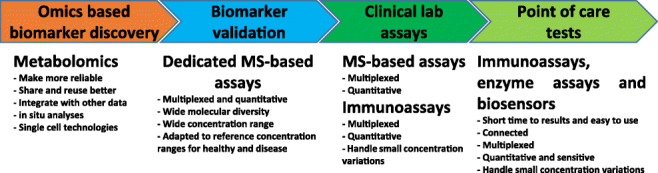
Metabolomics refers to the large-scale detection, quantification, and analysis of small molecules (metabolites) in biological media. Although metabolomics, alone or combined with other omics data, has already demonstrated its relevance for patient stratification in the frame of research projects and clinical studies, much remains to be done to move this approach to the clinical practice. This is especially true in the perspective of being applied to personalized/precision medicine, which aims at stratifying patients according to their risk of developing diseases, and tailoring medical treatments of patients according to individual characteristics in order to improve their efficacy and limit their toxicity. In this review article, we discuss the main challenges linked to analytical chemistry that need to be addressed to foster the implementation of metabolomics in the clinics and the use of the data produced by this approach in personalized medicine. First of all, there are already well-known issues related to untargeted metabolomics workflows at the levels of data production (lack of standardization), metabolite identification (small proportion of annotated features and identified metabolites), and data processing (from automatic detection of features to multi-omic data integration) that hamper the inter-operability and reusability of metabolomics data. Furthermore, the outputs of metabolomics workflows are complex molecular signatures of few tens of metabolites, often with small abundance variations, and obtained with expensive laboratory equipment. It is thus necessary to simplify these molecular signatures so that they can be produced and used in the field. This last point, which is still poorly addressed by the metabolomics community, may be crucial in a near future with the increased availability of molecular signatures of medical relevance and the increased societal demand for participatory medicine.
Related collections
Most cited references195

- Record: found
- Abstract: found
- Article: found
MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis
- Record: found
- Abstract: found
- Article: not found
Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking.
- Record: found
- Abstract: not found
- Article: not found