- Record: found
- Abstract: found
- Article: not found
Catalases Are NAD(P)H-Dependent Tellurite Reductases
Read this article at
Abstract
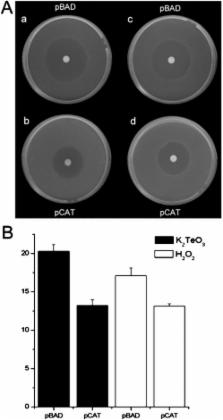
Reactive oxygen species damage intracellular targets and are implicated in cancer, genetic disease, mutagenesis, and aging. Catalases are among the key enzymatic defenses against one of the most physiologically abundant reactive oxygen species, hydrogen peroxide. The well-studied, heme-dependent catalases accelerate the rate of the dismutation of peroxide to molecular oxygen and water with near kinetic perfection. Many catalases also bind the cofactors NADPH and NADH tenaciously, but, surprisingly, NAD(P)H is not required for their dismutase activity. Although NAD(P)H protects bovine catalase against oxidative damage by its peroxide substrate, the catalytic role of the nicotinamide cofactor in the function of this enzyme has remained a biochemical mystery to date. Anions formed by heavy metal oxides are among the most highly reactive, natural oxidizing agents. Here, we show that a natural isolate of Staphylococcus epidermidis resistant to tellurite detoxifies this anion thanks to a novel activity of its catalase, and that a subset of both bacterial and mammalian catalases carry out the NAD(P)H-dependent reduction of soluble tellurite ion (TeO 3 2−) to the less toxic, insoluble metal, tellurium (Te°), in vitro. An Escherichia coli mutant defective in the KatG catalase/peroxidase is sensitive to tellurite, and expression of the S. epidermidis catalase gene in a heterologous E. coli host confers increased resistance to tellurite as well as to hydrogen peroxide in vivo, arguing that S. epidermidis catalase provides a physiological line of defense against both of these strong oxidizing agents. Kinetic studies reveal that bovine catalase reduces tellurite with a low Michaelis-Menten constant, a result suggesting that tellurite is among the natural substrates of this enzyme. The reduction of tellurite by bovine catalase occurs at the expense of producing the highly reactive superoxide radical.
Related collections
Most cited references53
- Record: found
- Abstract: not found
- Article: not found
Use of T7 RNA polymerase to direct expression of cloned genes.
- Record: found
- Abstract: found
- Article: not found
The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis.
- Record: found
- Abstract: found
- Article: not found
