- Record: found
- Abstract: found
- Article: found
Imbalance between Expression of FOXC2 and Its lncRNA in Lymphedema-Distichiasis Caused by Frameshift Mutations
Read this article at
Abstract
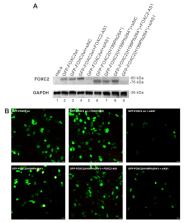
Forkhead-box C2 (FOXC2) is a transcription factor involved in lymphatic system development. FOXC2 mutations cause Lymphedema-distichiasis syndrome (LD). Recently, a natural antisense was identified, called lncRNA FOXC2-AS1, which increases FOXC2 mRNA stability. No studies have evaluated FOXC2 and FOXC2-AS1 blood expression in LD and healthy subjects. Here, we show that FOXC2 and FOXC-AS1 expression levels were similar in both controls and patients, and a significantly higher amount of both RNAs was observed in females. A positive correlation between FOXC2 and FOXC2-AS1 expression was found in both controls and patients, excluding those with frameshift mutations. In these patients, the FOXC2-AS1/ FOXC2 ratio was about 1:1, while it was higher in controls and patients carrying other types of mutations. The overexpression or silencing of FOXC2-AS1 determined a significant increase or reduction in FOXC2 wild-type and frameshift mutant proteins, respectively. Moreover, confocal and bioinformatic analysis revealed that these variations caused the formation of nuclear proteins aggregates also involving DNA. In conclusion, patients with frameshift mutations presented lower values of the FOXC2-AS1/ FOXC2 ratio, due to a decrease in FOXC2-AS1 expression. The imbalance between FOXC2 mRNA and its lncRNA could represent a molecular mechanism to reduce the amount of FOXC2 misfolded proteins, protecting cells from damage.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Forkhead box proteins: tuning forks for transcriptional harmony.
- Record: found
- Abstract: found
- Article: not found
The long noncoding RNA Malat1: Its physiological and pathophysiological functions.
- Record: found
- Abstract: found
- Article: not found