- Record: found
- Abstract: found
- Article: found
Claudin-2 Knockout by TALEN-Mediated Gene Targeting in MDCK Cells: Claudin-2 Independently Determines the Leaky Property of Tight Junctions in MDCK Cells
Read this article at
Abstract
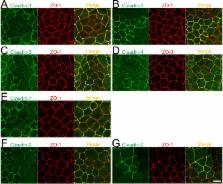
Tight junctions (TJs) regulate the movements of substances through the paracellular pathway, and claudins are major determinants of TJ permeability. Claudin-2 forms high conductive cation pores in TJs. The suppression of claudin-2 expression by RNA interference in Madin-Darby canine kidney (MDCK) II cells (a low-resistance strain of MDCK cells) was shown to induce a three-fold increase in transepithelial electrical resistance (TER), which, however, was still lower than in high-resistance strains of MDCK cells. Because RNA interference-mediated knockdown is not complete and only reduces gene function, we considered the possibility that the remaining claudin-2 expression in the knockdown study caused the lower TER in claudin-2 knockdown cells. Therefore, we investigated the effects of claudin-2 knockout in MDCK II cells by establishing claudin-2 knockout clones using transcription activator-like effector nucleases (TALENs), a recently developed genome editing method for gene knockout. Surprisingly, claudin-2 knockout increased TER by more than 50-fold in MDCK II cells, and TER values in these cells (3000–4000 Ω·cm 2) were comparable to those in the high-resistance strains of MDCK cells. Claudin-2 re-expression restored the TER of claudin-2 knockout cells dependent upon claudin-2 protein levels. In addition, we investigated the localization of claudin-1, -2, -3, -4, and -7 at TJs between control MDCK cells and their respective knockout cells using their TALENs. Claudin-2 and -7 were less efficiently localized at TJs between control and their knockout cells. Our results indicate that claudin-2 independently determines the ‘leaky’ property of TJs in MDCK II cells and suggest the importance of knockout analysis in cultured cells.
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Targeting DNA double-strand breaks with TAL effector nucleases.
- Record: found
- Abstract: found
- Article: not found
Claudin-1 and -2: Novel Integral Membrane Proteins Localizing at Tight Junctions with No Sequence Similarity to Occludin
- Record: found
- Abstract: found
- Article: not found