- Record: found
- Abstract: found
- Article: found
The rhizosphere microbiome of 51 potato cultivars with diverse plant growth characteristics
Read this article at
Abstract
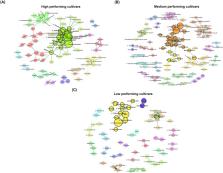
Rhizosphere microbial communities play a substantial role in plant productivity. We studied the rhizosphere bacteria and fungi of 51 distinct potato cultivars grown under similar greenhouse conditions using a metabarcoding approach. As expected, individual cultivars were the most important determining factor of the rhizosphere microbial composition; however, differences were also obtained when grouping cultivars according to their growth characteristics. We showed that plant growth characteristics were related to deterministic and stochastic assembly processes of bacterial and fungal communities, respectively. The bacterial genera Arthrobacter and Massilia (known to produce indole acetic acid and siderophores) exhibited greater relative abundance in high- and medium-performing cultivars. Bacterial co-occurrence networks were larger in the rhizosphere of these cultivars and were characterized by a distinctive combination of plant beneficial Proteobacteria and Actinobacteria along with a module of diazotrophs namely Azospira, Azoarcus, and Azohydromonas. Conversely, the network within low-performing cultivars revealed the lowest nodes, hub taxa, edges density, robustness, and the highest average path length resulting in reduced microbial associations, which may potentially limit their effectiveness in promoting plant growth. Our findings established a clear pattern between plant productivity and the rhizosphere microbiome composition and structure for the investigated potato cultivars, offering insights for future management practices.
Abstract
The study on 51 potato cultivars revealed a clear pattern between plant productivity and rhizosphere microbial composition, highlighting distinct features in high-performing cultivars conducive to enhanced plant growth.
Related collections
Most cited references93
- Record: found
- Abstract: found
- Article: not found
Modularity and community structure in networks
- Record: found
- Abstract: found
- Article: not found