- Record: found
- Abstract: found
- Article: found
Early immune factors associated with the development of post-acute sequelae of SARS-CoV-2 infection in hospitalized and non-hospitalized individuals

Read this article at
Abstract
Background
Infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can lead to post-acute sequelae of SARS-CoV-2 (PASC) that can persist for weeks to years following initial viral infection. Clinical manifestations of PASC are heterogeneous and often involve multiple organs. While many hypotheses have been made on the mechanisms of PASC and its associated symptoms, the acute biological drivers of PASC are still unknown.
Methods
We enrolled 494 patients with COVID-19 at their initial presentation to a hospital or clinic and followed them longitudinally to determine their development of PASC. From 341 patients, we conducted multi-omic profiling on peripheral blood samples collected shortly after study enrollment to investigate early immune signatures associated with the development of PASC.
Results
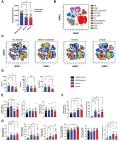
During the first week of COVID-19, we observed a large number of differences in the immune profile of individuals who were hospitalized for COVID-19 compared to those individuals with COVID-19 who were not hospitalized. Differences between individuals who did or did not later develop PASC were, in comparison, more limited, but included significant differences in autoantibodies and in epigenetic and transcriptional signatures in double-negative 1 B cells, in particular.
Conclusions
We found that early immune indicators of incident PASC were nuanced, with significant molecular signals manifesting predominantly in double-negative B cells, compared with the robust differences associated with hospitalization during acute COVID-19. The emerging acute differences in B cell phenotypes, especially in double-negative 1 B cells, in PASC patients highlight a potentially important role of these cells in the development of PASC.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles
- Record: found
- Abstract: found
- Article: not found
Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19
- Record: found
- Abstract: found
- Article: found