- Record: found
- Abstract: found
- Article: found
Replication Kinetics of B.1.351 and B.1.1.7 SARS-CoV-2 Variants of Concern Including Assessment of a B.1.1.7 Mutant Carrying a Defective ORF7a Gene
Read this article at
Abstract
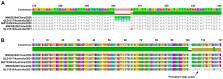
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19, is a readily transmissible and potentially deadly pathogen which is currently re-defining human susceptibility to pandemic viruses in the modern world. The recent emergence of several genetically distinct descendants known as variants of concern (VOCs) is further challenging public health disease management, due to increased rates of virus transmission and potential constraints on vaccine effectiveness. We report the isolation of SARS-CoV-2 VOCs imported into Australia belonging to the B.1.351 lineage, first described in the Republic of South Africa (RSA), and the B.1.1.7 lineage originally reported in the United Kingdom, and directly compare the replication kinetics of these two VOCs in Vero E6 cells. In this analysis, we also investigated a B.1.1.7 VOC (QLD1516/2021) carrying a 7-nucleotide deletion in the open reading frame 7a (ORF7a) gene, likely truncating and rendering the ORF7a protein of this virus defective. We demonstrate that the replication of the B.1.351 VOC (QLD1520/2020) in Vero E6 cells can be detected earlier than the B.1.1.7 VOCs (QLD1516/2021 and QLD1517/2021), before peaking at 48 h post infection (p.i.), with significantly higher levels of virus progeny. Whilst replication of the ORF7a defective isolate QLD1516/2021 was delayed longer than the other viruses, slightly more viral progeny was produced by the mutant compared to the unmutated isolate QLD1517/2021 at 72 h p.i. Collectively, these findings contribute to our understanding of SARS-CoV-2 replication and evolutionary dynamics, which have important implications in the development of future vaccination, antiviral therapies, and epidemiological control strategies for COVID-19.
Related collections
Most cited references24

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data
- Record: found
- Abstract: found
- Article: found