- Record: found
- Abstract: found
- Article: found
Application of Chest CT Imaging Feature Model in Distinguishing Squamous Cell Carcinoma and Adenocarcinoma of the Lung
Read this article at
Abstract
Purpose
In situations where pathological acquisition is difficult, there is a lack of consensus on distinguishing between adenocarcinoma and squamous cell carcinoma from imaging images, and each doctor can only make judgments based on their own experience. This study aims to extract imaging features of chest CT, extract sensitive factors through logistic univariate and multivariate analysis, and model to distinguish between lung squamous cell carcinoma and lung adenocarcinoma.
Methods
We downloaded chest CT scans with clear diagnosis of adenocarcinoma and squamous cell carcinoma from The Cancer Imaging Archive (TCIA), extracted 19 imaging features by a radiologist and a thoracic surgeon, including location, spicule, lobulation, cavity, vacuolar sign, necrosis, pleural traction sign, vascular bundle sign, air bronchogram sign, calcification, enhancement degree, distance from pulmonary hilum, atelectasis, pulmonary hilum and bronchial lymph nodes, mediastinal lymph nodes, interlobular septal thickening, pulmonary metastasis, adjacent structures invasion, pleural effusion. Firstly, we apply the glm function of R language to perform logistic univariate analysis on all variables to select variables with P < 0.1. Then, perform logistic multivariate analysis on the selected variables to obtain a predictive model. Next, use the roc function in R language to calculate the AUC value and draw the ROC curve, use the val.prob function in R language to draw the Calibrat curve, and use the rmda package in R language to draw the DCA curve and clinical impact curve. At the same time, 45 patients diagnosed with lung squamous cell carcinoma and lung adenocarcinoma through surgery or biopsy in the Radiotherapy Department and Thoracic Surgery Department of our hospital from 2023 to 2024 were included in the validation group. The chest CT features were jointly determined and recorded by the two doctors mentioned above and included in the validation group. The included image feature data are complete and does not require preprocessing, so directly entering statistical calculations. Perform ROC curves, calibration curves, DCA, and clinical impact curves in the validation group to further validate the predictive model. If the predictive model performs well in the validation group, further draw a nomogram to demonstrate.
Results
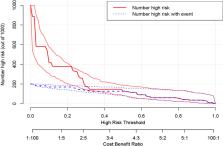
This study extracted 19 imaging features from the chest CT scans of 75 patients downloaded from TCIA and finally selected 18 complete data for analysis. First, univariate analysis and multivariate analysis were performed, and a total of 5 variables were obtained: spicule, necrosis, air bronchogram Sign, atelectasis, pulmonary hilum and bronchial lymph nodes. After conducting modeling analysis with AUC = 0.887, a validation group was established using clinical cases from our hospital, Draw ROC curve with AUC = 0.865 in the validation group, evaluate the accuracy of the model through Calibrate calibration curve, evaluate the reliability of the model in clinical practice through DCA curve, and further evaluate the practicality of the model in clinical practice through clinical impact curve.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer.

- Record: found
- Abstract: found
- Article: found
Prediction of pathologic stage in non-small cell lung cancer using machine learning algorithm based on CT image feature analysis

- Record: found
- Abstract: found
- Article: found