- Record: found
- Abstract: found
- Article: found
Network pharmacology evaluation of the active ingredients and potential targets of XiaoLuoWan for application to uterine fibroids
Read this article at
Abstract
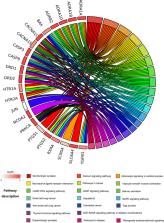
XiaoLuoWan (XLW) is a classical formula in traditional Chinese medicine (TCM) that has satisfactory therapeutic effects for uterine fibroids (UFs). However, its underlying mechanisms remain unclear. To elucidate the pharmacological actions of XLW in treating UFs, an ingredient–target–disease framework was proposed based on network pharmacology strategies. The active ingredients in XLW and their putative targets were obtained from the TCM systems pharmacology database and analysis platform (TCMSP) and Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine (BATMAN-TCM) platforms. The known therapeutic targets of UFs were acquired from the DigSee and DrugBank databases. Then, the links between putative XLW targets and therapeutic UF targets were identified to establish interaction networks by Cytoscape. Finally, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of overlapping gene targets were performed in the STRING database and visualized in R software. In total, 9 active compounds were obtained from 74 ingredients, with 71 curative targets predicted in XLW. Moreover, 321 known therapeutic targets were closely related to UFs, with 29 targets overlapping with XLW and considered interacting genes. Pathway enrichment revealed that the calcium signaling pathway was significantly enriched and the mitogen-activated protein kinase (MAPK) signaling pathway, cAMP signaling pathway, cancer and vascular smooth muscle contraction pathways, cGMP-PKG signaling pathway, and AGE-RAGE signaling pathway were closely associated with XLW intervention for UFs. In conclusion, the network pharmacology detection identified 9 available chemicals as the active ingredients in XLW that may relieve UFs by regulating 29 target genes involved in the calcium signaling pathway, MAPK pathway and cAMP pathway. Network pharmacology analyses may provide more convincing evidence for the investigation of classical TCM prescriptions, such as XLW.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
clusterProfiler: an R package for comparing biological themes among gene clusters.
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.

- Record: found
- Abstract: found
- Article: found