- Record: found
- Abstract: found
- Article: not found
C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance
Summary:
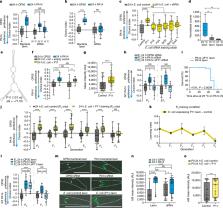
C. elegans must distinguish pathogenic from nutritious bacterial food sources among the many bacteria it is exposed to in its environment 1 . Here we show that a single exposure to purified small RNAs isolated from pathogenic Pseudomonas aeruginosa (PA14) is sufficient to induce pathogen avoidance, both in the treated animals and in four subsequent generations of progeny. The RNA interference and piRNA pathways, the germline, and the ASI neuron are required for bacterial small RNA-induced avoidance behavior and transgenerational inheritance. A single P. aeruginosa non-coding RNA, P11, is both necessary and sufficient to convey learned avoidance of PA14, and its C. elegans target, maco-1, is required for avoidance. Our results suggest that this ncRNA-dependent mechanism evolved to survey the worm’s microbial environment, use this information to make appropriate behavioral decisions, and pass this information on to its progeny.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Mfold web server for nucleic acid folding and hybridization prediction.
- Record: found
- Abstract: found
- Article: not found
Small silencing RNAs: an expanding universe.
- Record: found
- Abstract: not found
- Article: not found
Moving beyond P values: data analysis with estimation graphics
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.