- Record: found
- Abstract: found
- Article: found
FLI1 is associated with regulation of DNA methylation and megakaryocytic differentiation in FPDMM caused by a RUNX1 transactivation domain mutation
Read this article at
Abstract
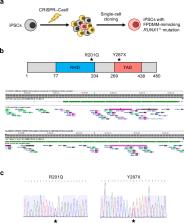
Familial platelet disorder with associated myeloid malignancies (FPDMM) is an autosomal dominant disease caused by heterozygous germline mutations in RUNX1. It is characterized by thrombocytopenia, platelet dysfunction, and a predisposition to hematological malignancies. Although FPDMM is a precursor for diseases involving abnormal DNA methylation, the DNA methylation status in FPDMM remains unknown, largely due to a lack of animal models and challenges in obtaining patient-derived samples. Here, using genome editing techniques, we established two lines of human induced pluripotent stem cells (iPSCs) with different FPDMM-mimicking heterozygous RUNX1 mutations. These iPSCs showed defective differentiation of hematopoietic progenitor cells (HPCs) and megakaryocytes (Mks), consistent with FPDMM. The FPDMM-mimicking HPCs showed DNA methylation patterns distinct from those of wild-type HPCs, with hypermethylated regions showing the enrichment of ETS transcription factor (TF) motifs. We found that the expression of FLI1, an ETS family member, was significantly downregulated in FPDMM-mimicking HPCs with a RUNX1 transactivation domain (TAD) mutation. We demonstrated that FLI1 promoted binding-site-directed DNA demethylation, and that overexpression of FLI1 restored their megakaryocytic differentiation efficiency and hypermethylation status. These findings suggest that FLI1 plays a crucial role in regulating DNA methylation and correcting defective megakaryocytic differentiation in FPDMM-mimicking HPCs with a RUNX1 TAD mutation.
Related collections
Most cited references68
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools
- Record: found
- Abstract: found
- Article: not found