- Record: found
- Abstract: found
- Article: found
Biocontrol of Bacterial Leaf Blight of Rice and Profiling of Secondary Metabolites Produced by Rhizospheric Pseudomonas aeruginosa BRp3
Read this article at
Abstract
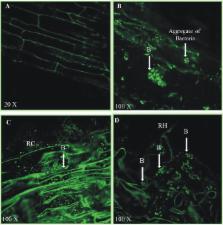
Xanthomonas oryzae pv. oryzae (Xoo) is widely prevalent and causes Bacterial Leaf Blight (BLB) in Basmati rice grown in different areas of Pakistan. There is a need to use environmentally safe approaches to overcome the loss of grain yield in rice due to this disease. The present study aimed to develop inocula, based on native antagonistic bacteria for biocontrol of BLB and to increase the yield of Super Basmati rice variety. Out of 512 bacteria isolated from the rice rhizosphere and screened for plant growth promoting determinants, the isolate BRp3 was found to be the best as it solubilized 97 μg/ mL phosphorus, produced 30 μg/mL phytohormone indole acetic acid and 15 mg/ L siderophores in vitro. The isolate BRp3 was found to be a Pseudomonas aeruginosa based on 16S rRNA gene sequencing (accession no. HQ840693). This bacterium showed antagonism in vitro against different phytopathogens including Xoo and Fusarium spp. Strain BRp3 showed consistent pathogen suppression of different strains of BLB pathogen in rice. Mass spectrometric analysis detected the production of siderophores (1-hydroxy-phenazine, pyocyanin, and pyochellin), rhamnolipids and a series of already characterized 4-hydroxy-2-alkylquinolines (HAQs) as well as novel 2,3,4-trihydroxy-2-alkylquinolines and 1,2,3,4-tetrahydroxy-2-alkylquinolines in crude extract of BRp3. These secondary metabolites might be responsible for the profound antibacterial activity of BRp3 against Xoo pathogen. Another contributing factor toward the suppression of the pathogen was the induction of defense related enzymes in the rice plant by the inoculated strain BRp3. When used as an inoculant in a field trial, this strain enhanced the grain and straw yields by 51 and 55%, respectively, over non-inoculated control. Confocal Laser Scanning Microscopy (CLSM) used in combination with immunofluorescence marker confirmed P. aeruginosa BRp3 in the rice rhizosphere under sterilized as well as field conditions. The results provide evidence that novel secondary metabolites produced by BRp3 may contribute to its activity as a biological control agent against Xoo and its potential to promote the growth and yield of Super Basmati rice.
Related collections
Most cited references97
- Record: found
- Abstract: not found
- Article: not found
Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium
- Record: found
- Abstract: found
- Article: not found
Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years.
- Record: found
- Abstract: found
- Article: not found