- Record: found
- Abstract: found
- Article: found
A genome-wide association study of serum proteins reveals shared loci with common diseases
Read this article at
Abstract
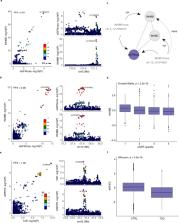
With the growing number of genetic association studies, the genotype-phenotype atlas has become increasingly more complex, yet the functional consequences of most disease associated alleles is not understood. The measurement of protein level variation in solid tissues and biofluids integrated with genetic variants offers a path to deeper functional insights. Here we present a large-scale proteogenomic study in 5,368 individuals, revealing 4,035 independent associations between genetic variants and 2,091 serum proteins, of which 36% are previously unreported. The majority of both cis- and trans-acting genetic signals are unique for a single protein, although our results also highlight numerous highly pleiotropic genetic effects on protein levels and demonstrate that a protein’s genetic association profile reflects certain characteristics of the protein, including its location in protein networks, tissue specificity and intolerance to loss of function mutations. Integrating protein measurements with deep phenotyping of the cohort, we observe substantial enrichment of phenotype associations for serum proteins regulated by established GWAS loci, and offer new insights into the interplay between genetics, serum protein levels and complex disease.
Abstract
Circulating proteins have been linked to many conditions, and understanding their genetic control can lead to understanding of disease. Here, the authors associate common genetic variants with protein levels, finding overlap of genetic associations with circulating proteins and complex disease.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
Proteomics. Tissue-based map of the human proteome.

- Record: found
- Abstract: found
- Article: found
Second-generation PLINK: rising to the challenge of larger and richer datasets
- Record: found
- Abstract: found
- Article: found